Abstract
Proteasome-dependent turnover of mitochondrial outer membrane (OMM)-associated proteins is one of the mechanisms for maintaining proper mitochondrial quality and function. However, the underlying pathways and their implications in human disease are poorly understood. Huntington’s disease (HD) is a fatal, inherited neurodegenerative disorder caused by expanded CAG repeats in the N terminal of the huntingtin gene (mutant Huntingtin, mtHtt). In this study, we show an extensive degradation of the OMM protein MCL1 (Myeloid cell leukemia sequence 1) in both HD mouse striatal cells and HD patient fibroblasts. The decrease in MCL1 level is associated with mitochondrial and cellular damage. Valosin-containing-protein (VCP) is an AAA-ATPase central to protein turnover via the ubiquitin proteasome system (UPS). We found that VCP translocates to mitochondria and promotes MCL1 degradation in HD cell cultures. Either down-regulation of VCP by RNA interference or inhibition of VCP by a dominant negative mutant abolishes MCL1 degradation in HD cell cultures. We further show that UBX-domain containing protein 1 (UBXD1), a known co-factor of VCP assisting in the recognition of substrates for protein degradation, selectively binds to MCL1 and interacts with VCP to mediate MCL1 extraction from the mitochondria. These results indicate that the OMM protein MCL1 is degraded by the VCP-UBXD1 complex and that the process is promoted by the presence of mtHtt. Therefore, our finding provides a new insight into the mechanism of mitochondrial dysfunction in HD.
Introduction
Huntington’s disease (HD) is a fatal, inherited neurodegenerative disorder that progresses for 15–20 years after diagnosis. The mutation that causes HD is a variable expansion of CAG repeats encoding polyglutamine (polyQ) in the huntingtin (Htt) protein (mutant Huntingtin, mtHtt) [1]. Many studies, including ours, show that mitochondrial dysfunction is one of the major events in the pathogenesis of HD [2–6]. Mitochondria isolated from HD patients or HD-transgenic mouse brains show an increase in mitochondrial permeability, perturbation in mitochondrial membrane potential, changes in mitochondrial ultrastructure and DNA integrity, as well as failure in bio-energetics [7–10]. Moreover, incubation of isolated mitochondria with recombinant mtHtt protein leads to mitochondrial swelling and release of cytochrome c [10, 11], suggesting direct mitochondrial injury by mtHtt. Further, we and others recently demonstrated the impairment of mitochondrial fission and mitophagy in various experimental models of HD [3, 5, 6, 12, 13] and that pharmacological inhibition of either mitochondrial fragmentation or aberrant mitophagy reduces neuropathological and behavioral phenotypes in HD transgenic mice [5, 6]. Thus, proper control of mitochondrial quality is likely to be a useful strategy for halting or slowing the pathology of HD.
To ensure proper mitochondrial function, diverse mitochondrial quality control mechanisms are evolutionarily conserved. In addition to mitochondrial dynamics (fusion and fission) that are critical for maintaining mitochondrial morphology and transport [12, 14], mitochondria-associated protein degradation (MAD) that occurs at the compartment of the outer mitochondrial membrane (OMM) has been proposed to be one of the mechanisms for controlling normal mitochondrial function [15, 16]. The MAD mechanism involves retro-translocation of ubiquitinated proteins from the OMM to the cytosol where the proteins can be degraded by the ubiquitin-proteasome pathway (UPS) [15, 17, 18].
Valosin-containing-protein (VCP), also known as p97, is an AAA-ATPase central to UPS-dependent protein turnover [19]. VCP uses energy derived from ATP hydrolysis to apply mechanical force to substrates, thereby extracting substrates from diverse cellular locations for proteasomal degradation [19, 20]. Several studies reported that VCP translocates to the mitochondria, where it targets ubiquitinated OMM proteins (substrates) with relatively short half-lives before retro-translocating them to the proteasome for degradation [21–24]. We recently found that VCP is recruited to the mitochondria in various experimental models of HD and that blocking VCP mitochondrial translocation by a novel inhibitor reduces neuropathology of HD [5], indicating the importance of mitochondrial VCP in HD. However, whether mitochondria-accumulated VCP is involved in the MAD mechanism in HD is unknown.
Myeloid cell leukemia sequence 1 (MCL1) is a nucleus-encoded protein localized both on the OMM and in the mitochondrial matrix [5]. MCL1 is essential for maintaining mitochondrial membrane potential and is required for matrix and inner membrane structure integrity and mitochondrial bioenergetics [25, 26]. MCL1 can also serve as a stress sensor that regulates autophagy [27]. The protein level of MCL1 is controlled by the UPS. Degradation of MCL1 in cells results in extensive mitochondrial damage (membrane depolarization, increased oxidative stress, defects in bioenergetics, loss of cristae structure, and mitochondrial DNA depletion) [25–29], which ultimately leads to necrosis [27–29], apoptosis [30, 31], and augmented autophagy [26, 28, 29]. Thus, the stability of MCL1 is of great importance in maintaining overall mitochondrial quality and cell viability.
In this study, we observe MCL1 degradation in both cells expressing mtHtt and HD patient fibroblasts. Moreover, we identify that VCP and its co-factor UBXD1 mediate MCL1 degradation, the process of which is promoted by the presence of mtHtt. Thus, our study establishes an association of the MAD dysregulation with HD pathogenesis.
Results
MCL1 is degraded in HD cell cultures
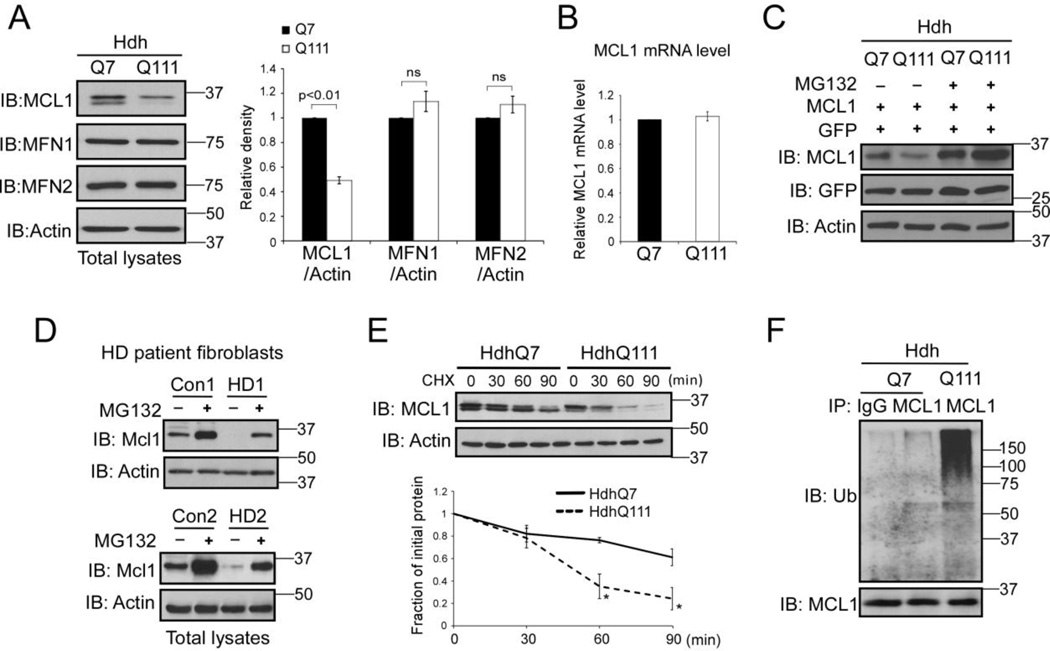
To determine if the MAD is disturbed in HD, we first investigated the protein levels of the OMM proteins in HdhQ7 and HdhQ111 knock-in mouse striatal cells. HdhQ7 and Q111 cells were immortalized from knock-in mice carrying 7 and 111 CAG repeats, respectively, in the mouse htt gene [32]. The HdhQ111 cells express a full-length mtHtt at endogenous level and therefore provide a genetically accurate cell model of HD [32]. Proteins measured included MCL1, MFN1, and MFN2, which are known substrates of the MAD [21–24]. Western blot analysis showed that the protein level of MCL1 was decreased in HdhQ111 mutant mouse striatal cells relative to that in HdhQ7 wildtype cells (Fig. 1A). However, there were no changes in the protein levels of MFN1 and MFN2 observed between HdhQ7 and HdhQ111 cells (Fig. 1A). By contrast, the mRNA level of MCL1 was not affected in HdhQ111 mutant cells (Fig. 1B). To determine whether the decrease in MCL1 protein levels in the HdhQ111 mutant cells is the result of protein degradation via the UPS, we over-expressed the MCL1 plasmid in HdhQ7 and HdhQ111 striatal cells in the presence or absence of MG132, an inhibitor of proteasome activity. We observed a great down-regulation of MCL1 in HdhQ111 cells, which was blocked by the addition of MG132 (Fig. 1C). Consistently, MG132 treatment abolished MCL1 degradation occurred in two lines of HD patient fibroblasts (Fig. 1D). To further test whether MCL1 stability is disturbed by mutant Htt (mtHtt), we performed a cycloheximide (CHX) chase assay in both HdhQ7 and HdhQ111 cells. Western blot analysis revealed that the half-life of MCL1 was markedly reduced in HdhQ111 cells compared to that in wildtype HdhQ7 cells (Fig. 1E). In addition, we observed an increase in poly-ubiquitination of MCL1 in HdhQ111 cells (Fig. 1F). Taken together, these results demonstrate that MCL1 was selectively degraded in HD cell cultures via the UPS pathway.
Figure 1. MCL1 is degraded in HD cell cultures via the UPS pathway.
(A) Total lysates of HdhQ7 and HdhQ111 mouse striatal cells were harvested. Protein levels of MCL1, MFN1, and MFN2 were determined by Western blot (WB) analysis using the indicated antibodies. Actin was used as a loading control. The data shown in the histogram are mean ± SE from three independent experiments. Student t-test. (B) RNA was extracted from HdhQ7 and HdhQ111 cells. The mRNA level of MCL1 was determined by real-time PCR. GAPDH was an internal control. The data are mean ± SE from three independent experiments. (C) HdhQ7 and HdhQ111 cells were co-transfected with MCL1 and GFP plasmids, followed by the addition of the proteasome inhibitor MG132 (5 µM for 16 hours). The MCL1 protein level was determined by WB. GFP was used as a control to ensure equal transfection efficiency among experimental groups. The shown blots are representative from three independent experiments. (D) HD patient fibroblasts (GM04693 and GM05539) and normal fibroblasts (Con1 and Con 2) were treated with or without MG132 (5 µM for 16 hours). Total lysates were subjected to WB analysis with the indicated antibodies. The shown blots are from three independent experiments. (E) Cycloheximide (CHX)-chase assay of MCL1 in HdhQ7 or HdhQ111 cells was conducted. The cells were treated with CHX at 50 µg/ml for the indicated time. The half-life of MCL1 was determined by WB. Actin was used as a loading control. The data are mean ± SE of three independent experiments. p<0.05, ANOVA with post-hoc Holm-Sidak test. (F) HdhQ7 and HdhQ111 cells were treated with MG132 (5 µM, 16 hours). Immunoprecipitation (IP) with anti-MCL1 antibody followed by WB with anti-ubiquitin (Ub) was performed. Note: the amount of protein used for IP in HdhQ111 cells was modified to ensure similar levels of MCL1 protein in each group (see input). The blots shown are from three independent experiments.
Decrease in MCL1 causes mitochondrial damage and enhances autophagy
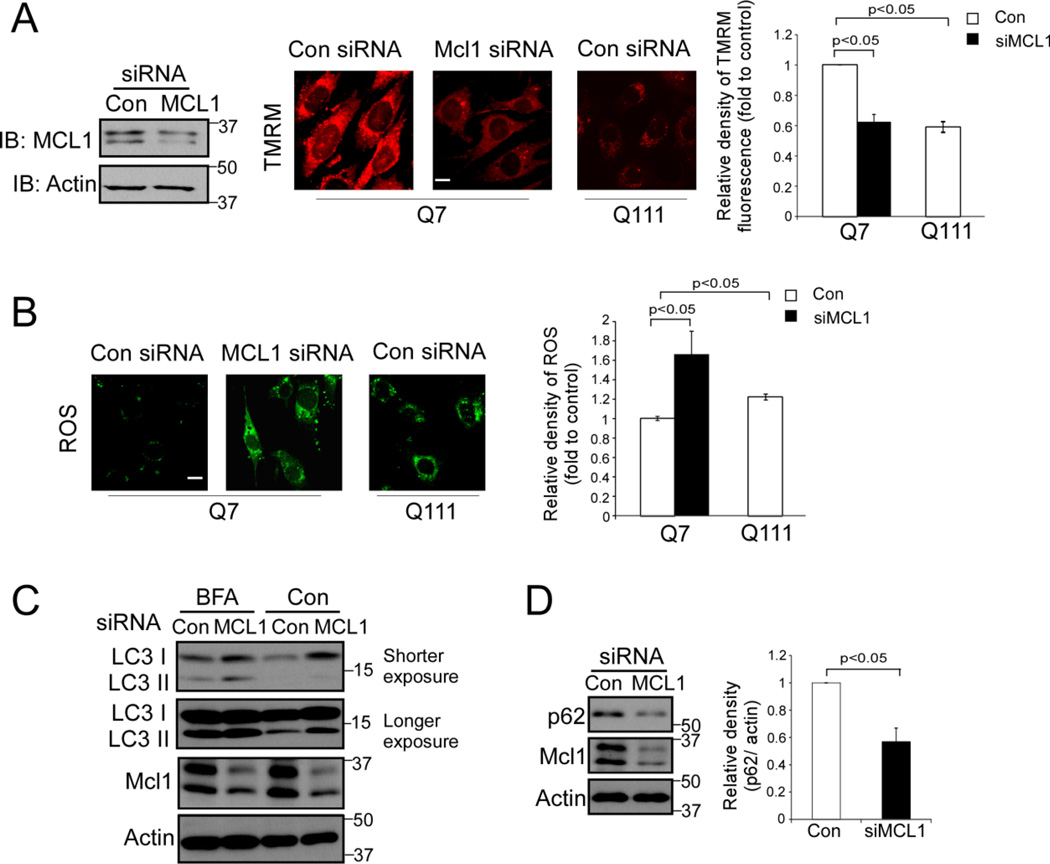
To determine the functional consequence of MCL1 degradation, we down-regulated MCL1 by small RNA interference (siRNA) and examined mitochondrial function and cellular survival. In wildtype HdhQ7 mouse striatal cells, knocking down MCL1 resulted in a decrease in mitochondrial membrane potential (Fig. 2A) and an increase in the production of reactive oxygen species (ROS) (Fig. 2B). Similarly, HdhQ111 mouse striatal cells in which the MCL1 protein level is low (Fig. 1A), exhibited lower mitochondrial membrane potential and higher ROS than those in HdhQ7 cells (Fig. 2A and 2B).
Figure 2. A decrease in MCL1 causes mitochondrial and cellular damage.
MCL1 was down-regulated by siRNA in HdhQ7 wildtype mouse striatal cells. (A) Left: Western blots confirmed the knock-down of MCL1. Middle: Mitochondrial membrane potential was determined using Tetramethylrhodamine methyl ester (TMRM) at the indicated groups. Scale bar: 10 µm. Right: quantitative analysis of TMRM fluorescence density. At least 100 cells per group are counted. (B) An ROS indicator was used to determine the production of reactive oxygen species (ROS). Left: representative images of ROS probes in cells at the indicated groups. Scale bar: 10 µm. Right: histogram of quantitative analysis. At least 100 cells per group are counted. (C) The LC3 protein level was determined by WB analysis in the presence or absence of BFA (20 nM for 4 hours). Shown blots are from three independent experiments. (D) Total protein level of p62 was determined by WB analysis. Histogram: quantitative analysis of total protein level of p62 vs. Actin. All the data are mean ± SE of three independent experiments. ANOVA with post-hoc Holm-Sidak test.
The conversion of microtubule-associated protein 1A/1B-light chain 3 (LC3) I into its lipidated form LC3II is an indicator of autophagy [33]. Increased levels of LC3II in the presence of Bafilomycin A1 (BFA, an inhibitor to prevent maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes [34]) has been used to evaluate autophagosome formation [35]. We found that silencing MCL1 in widltype mouse striatal cells increased the levels of LC3II in the presence of BFA. Further, down-regulation of MCL1 reduced the level of p62, one factor to indicate autophagy turnover [36]. These data suggest that decreased MCL1 lead to an increase in autophagy. Collectively, our findings in cultured cells are consistent with previous studies [25–29], showing that the decrease in MCL1 results in mitochondrial and cellular damage.
VCP is required for MCL1 degradation in HD cell cultures
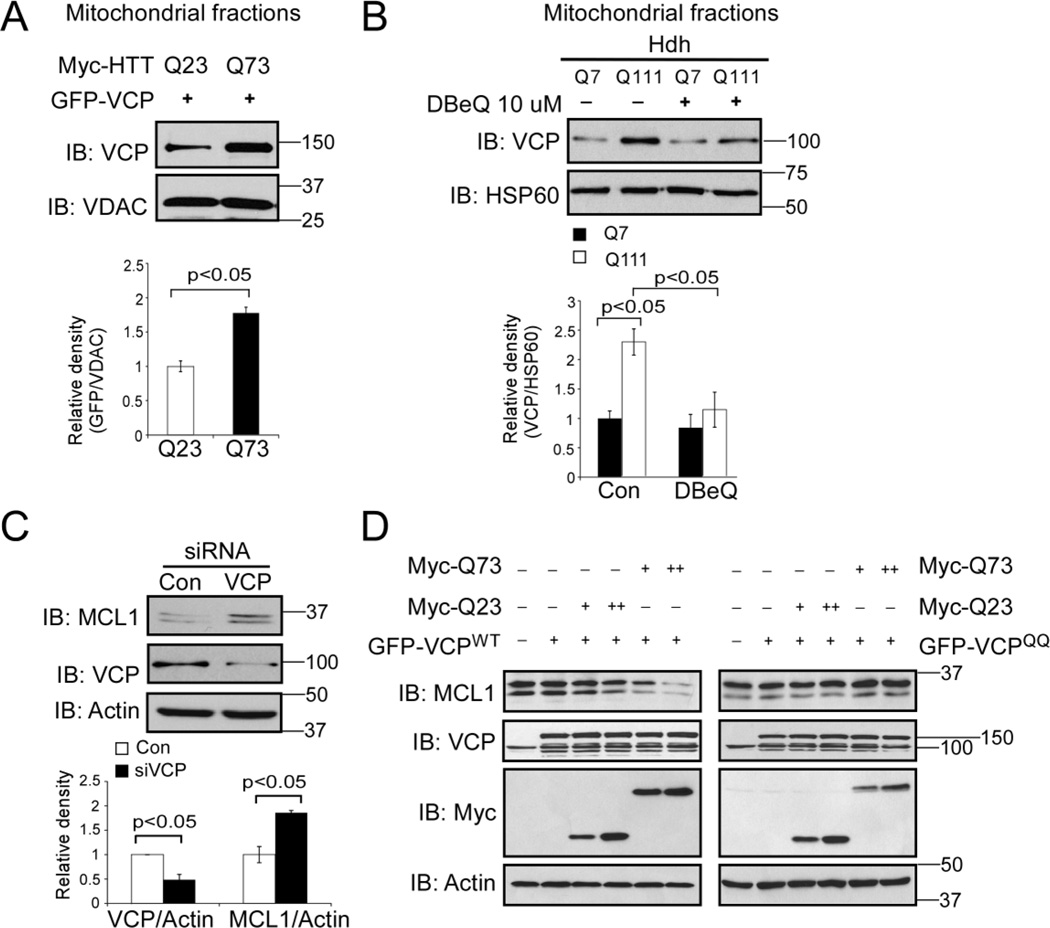
VCP regulates the turnover of MCL1 under normal and apoptotic stress conditions [24]. To examine whether VCP is required for MCL1 degradation in our HD cell cultures, we co-transfected GFP-VCP with Myc-tagged full-length Htt with 23 (Q23, wildtype) or 73 (Q73, mutant) CAG repeats in HEK293T cells. We found more GFP-VCP associated with the mitochondria of cells expressing Myc-Htt Q73 than that of Myc-Htt Q23-expressing cells (Fig. 3A), suggesting that mtHtt recruit VCP to the mitochondria. Consistent with this observation, the mitochondrial levels of VCP increased in HdhQ111 cells relative to those measured in HdhQ7 cells (Fig. 3B). Treatment with N2,N4-dibenzylquinazoline-2,4-diamine (DBeQ), a VCP inhibitor that inhibits VCP ATPase activity [37], abolished this VCP mitochondrial accumulation in the HdhQ111 cells (Fig. 3B). Furthermore, down-regulation of VCP by siRNA in HdhQ111 cells resulted in an increase in the protein level of MCL1 (Fig. 3C). A dominant negative VCP ATPase mutant (VCPE305Q,E578Q, VCPQQ) has been shown to inhibit VCP-mediated substrate retro-translocation from the cellular membrane to the cytosol and therefore stabilizes VCP substrates [38]. We co-transfected either GFP-VCPWT or its dominant negative mutant GFP-VCPQQ with Myc-tagged Htt Q23 or Q73 plasmids in HEK293T cells. As shown in Fig. 3D, we found that overexpression of Myc-Q73 promoted MCL1 degradation in cells expressing GFP-VCPWT, whereas mtHtt-induced MCL1 degradation was abolished in cells expressing the GFP-VCPQQ mutant. These data collectively suggest that mitochondria-accumulated VCP is required for MCL1 degradation in HD cell cultures.
Figure 3. VCP is required for MCL1 degradation in HD cell cultures.
(A) HEK293T cells were transfected with Myc-tagged, full-length Htt with 23 or 73 CAG repeats (Myc-Htt-23Q and Myc-Htt-73Q, respectively) for 36 hours. Mitochondrial fractions were isolated and WB analysis was performed with anti-VCP antibody. VDAC was used as a loading control. The data are mean ± SE of three independent experiments. Student t-test. (B) HdhQ7 and HdhQ111 cells were treated with VCP inhibitor DBeQ (10 µM) for 16 hours. Mitochondria were isolated and the VCP protein level was determined by WB analysis. HSP60 was a loading control. The data are mean ± SE of three independent experiments. ANOVA with post-hoc Holm-Sidak test. (C) HdhQ111 cells were transfected with control or VCP siRNA for 48 hours. MCL1 and VCP protein levels were examined by WB with anti-MCL1 and anti-VCP antibodies. The data are mean ± SE of three independent experiments. ANOVA with post-hoc Holm-Sidak test. (D) HEK293T cells were co-transfected GFP-VCP wildtype (GFP-VCPWT) or QQ mutant (GFP-VCPQQ) with Myc-tagged Htt carrying 23 (Myc-Q23) or 73 (Myc-73Q) CAG repeats for 36 hours. Western blot analysis was conducted with the indicated antibodies. The blots shown are from three independent experiments.
UBXD1 is a cofactor of VCP on the mitochondria
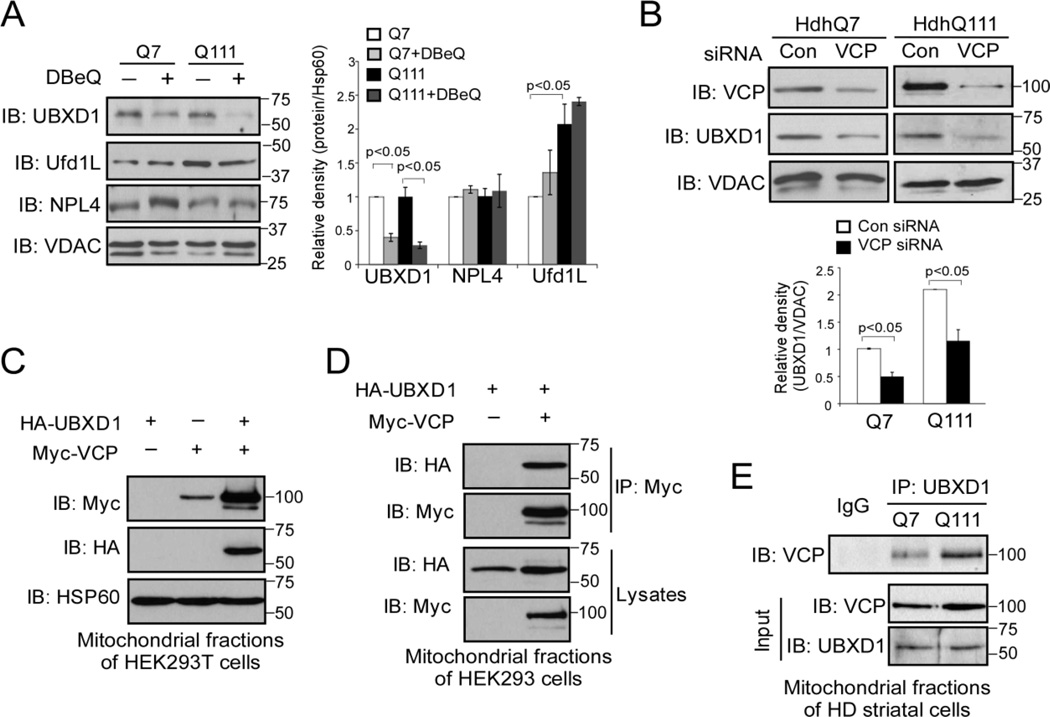
VCP functions in diversity of cellular processes, including ER-associated protein degradation (ERAD), mitochondria-associated degradation (MAD), autophagy and DNA repair by localizing at different subcellular organelles [19]. The coordination of these diverse cellular pathways is defined by VCP cofactors, a group of proteins containing a ubiquitin-X (UBX) domain or UBX-like domain [19, 39]. These domains assume a ubiquitin fold and interact with VCP to form a protein complex [39, 40]. The major cofactors of VCP include UFD1L (ubiquitin fusion degradation 1L), NPL4 (nuclear protein localization homolog 4) heterodimer, p47, and UBXD1 (UBX-domain containing protein 1) [40–43]. These major cofactors have been shown to modulate VCP activity or govern assembly of additional cofactors. Each core complex can then act in several pathways by associating with alternative sets of accessory proteins that determine localization of VCP [19, 39]. Because the association of VCP and its cofactor with mitochondria is required for the recognition of mitochondrial substrates, we hypothesize that disturbance of VCP’s association with mitochondria would affect the level of VCP mitochondrial co-factors associated with the organelle. We treated HdhQ7 and HdhQ111 cells with the VCP inhibitor DBeQ, which blocked VCP translocation to the mitochondria (Fig. 3B), and determined the levels of several known VCP co-factors on the mitochondrial fractions of these cells by Western blot analysis. We found that association of UBXD1 with the mitochondria was greatly inhibited by treatment with DBeQ in both HdhQ7 and HdhQ111 striatal cells (Fig. 4A). In contrast, treatment of DBeQ had no effects on the mitochondrial levels of NPL4 and UFD1L, although the UFD1L mitochondrial level was enhanced in HdhQ111 cells (Fig. 4A). Additionally, down-regulation of VCP in both HdhQ7 and HdhQ11 striatal cells by VCP siRNA reduced the level of UBXD1 on the mitochondria (Fig. 4B), whereas overexpression of Myc-VCP promoted HA-UBXD1 association with the mitochondria (Fig. 4C). Finally, we demonstrated the interaction between VCP and UBXD1 on the mitochondria of cells. As shown in Fig. 4D, HA-UBXD1 co-immunoprecipitated with Myc-VCP in HEK293T cells. A further co-immunoprecipitation assay revealed that the endogenous binding of VCP and UBXD1 on the mitochondria was stronger in HdhQ111 cells than in HdhQ7 cells (Fig. 4E). Thus, UBXD1 might be a cofactor of VCP on mitochondria under the conditions associated with HD.
Figure 4. UBXD1 is a VCP mitochondrial cofactor.
(A) HdhQ7 and HdhQ111 cells were treated with VCP inhibitor DBeQ (10 µM) for 16 hours. Western blot analysis of mitochondrial fractions was performed with the indicated antibodies. The data are mean ± SE of three independent experiments. *, p<0.05 vs. Q111 cells. ANOVA with post-hoc Holm-Sidak test. (B) HdhQ111 cells were transfected with control or VCP siRNA for 48 h. Mitochondrial VCP and UBXD1 protein levels were detected with anti-VCP and anti-UBXD1 antibodies by WB. The data are mean ± SE of three independent experiments. *, p<0.05 vs. cells with con siRNA. ANOVA with post-hoc Holm-Sidak test. (C) Control vector or Myc-VCP was co-expressed with HA-UBXD1 in HEK293T cells for 36 hours. Western blot analyses of mitochondrial fractions were performed with anti-Myc, anti-HA, and anti-HSP60 antibodies. (D) HEK293T cells were co-transfected with Myc-VCP and HA-UBXD1 for 36 hours. Immunoprecipitates (IP) with anti-Myc were immunoblotted (IB) with anti-HA antibody. (E) Mitochondrial fractions were isolated from HdhQ7 and HdhQ111 cells. IP with anti-UBXD1 followed by IB with anti-VCP antibody was conducted. All blots shown are from three independent experiments.
UBXD1 binds to MCL1 and cooperates with VCP to extract MCL1 from mitochondria in HD cell culture
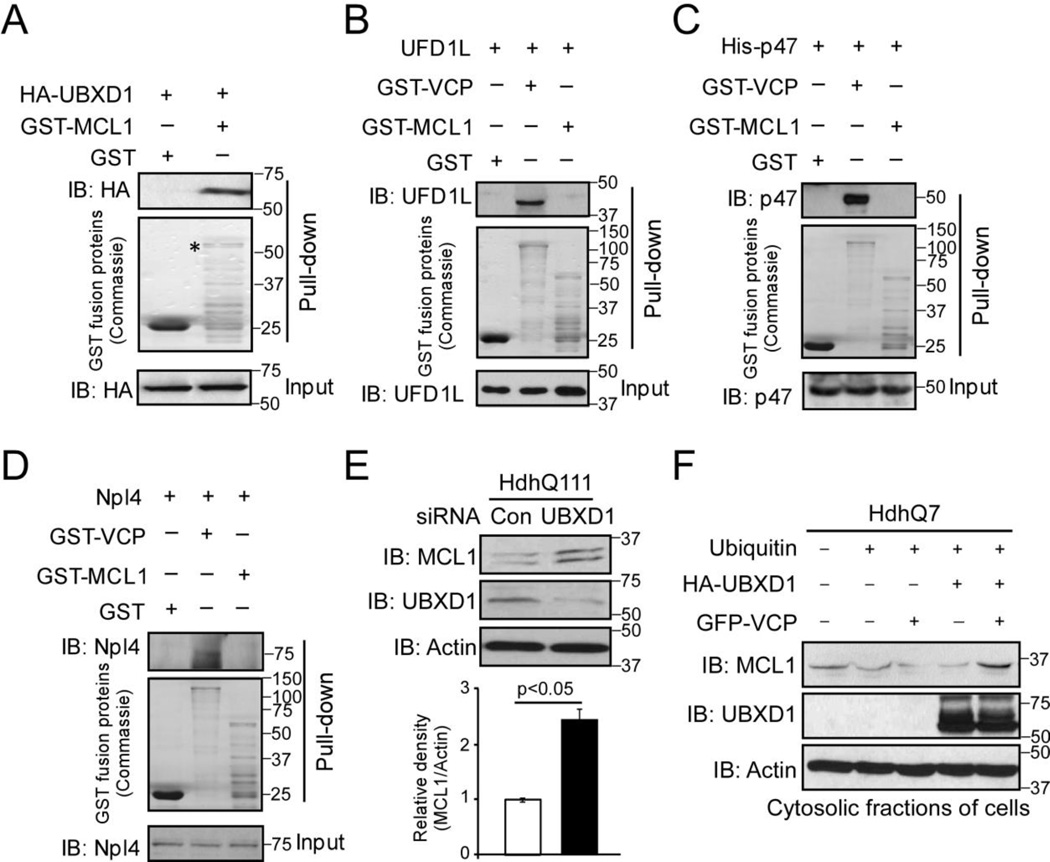
VCP substrates are bound to and extracted by the cofactors of VCP for degradation [44]. We conducted a GST pull-down assay to determine the interaction between UBXD1 and MCL1. We found that GST-MCL1 was bound to HA-UBXD1 in vitro (Fig. 5A). In contrast, there was no interaction between MCL1 and VCP cofactors, including UFD1L, p47, and NPL4, in the GST pull-down assay, even though VCP did bind to these co-factors (Fig. 5B, C, D). Furthermore, in HdhQ111 mutant striatal cells, down-regulation of UBXD1 by siRNA elevated the protein level of MCL1 (Fig. 5E). These findings support our hypothesis that UBXD1 facilitates VCP-mediated MCL1 degradation in HD. Next, we examined the role of UBXD1 in MCL1 retro-translocation from the OMM to the cytosolic fractions, an essential step to process UPS-dependent degradation. In wildtype mouse striatal cells, we expressed GFP-VCP and HA-UBXD1 in the presence or absence of ubiquitin. We then isolated cytosolic fractions and determined the level of MCL1. Expression of either GFP-VCP or HA-UBXD1 alone did not affect the protein level of MCL1 in the cytosolic fractions. By contrast, co-expression of VCP and UBXD1 promoted the accumulation of MCL1 in the cytosolic fractions (Fig. 5F). These results suggest that VCP and UBXD1 work interdependently to extract MCL1 from the mitochondria, therefore promoting MCL1 retro-translocation and degradation.
Figure 5. UBXD1 and VCP cooperate to extract MCL from the mitochondria.
(A) HA-UBXD1 was transfected in HdhQ7 wildtype striatal cells for 36 hours. Total cell lysates were incubated with GST or GST-MCL1 overnight. Immunoprecipitates (IP) were analyzed by IB with anti-HA antibody. Total cell lysates of HdhQ7 wildtype cells were incubated overnight with GST, GST-MCL1, or GST-VCP. Immunoprecipitates were analyzed by IB with anti-UFD1L (B) or an-NPL4 (D) antibodies. (C) Bacteria-expressed His-p47 was incubated overnight with GST, GST-MCL1, or GST-VCP. Immunoprecipitates were analyzed by IB with p47 antibody. (E) HdhQ111 mutant striatal cells were transfected with control siRNA or UBXD1 siRNA for 48 hours. MCL1 and UBXD1 were detected by WB with the indicated antibodies. Actin was used as a loading control. (F) Ubiquitin was co-transfected with HA-UBXD1 and GFP-VCP in HdhQ7 wildtype cells for 48 hours. Cytosolic fractions were isolated and analyzed by IB with anti-MCL1 and anti-UBXD1 antibodies. All blots shown are from three independent experiments.
Discussion
The OMM separates mitochondria from the cytosol and are important for maintaining mitochondrial integrity, including regulation of apoptosis, mitochondrial membrane fusion and fission as well as mitophagy [16]. Findings from multiple groups suggest a link between the MAD pathway that targets proteins at the OMM for protein turnover and mitochondrial quality control [21, 23, 24]. However, whether the mechanism of the MAD is implicated in human diseases is not yet explored. In this study, using the cell culture models of HD, we reported for the first time that the MAD pathway is disturbed in HD; mtHtt causes extensive protein degradation of MCL1, an OMM protein required for mitochondrial membrane integrity and autophagy. Moreover, we found that mtHtt recruits VCP to the mitochondria, where VCP and UBXD1 form a protein complex to mediate MCL1 protein degradation. Therefore, our findings suggest a novel mechanism by which failure in mitochondrial protein quality control contributes to the pathogenesis of HD.
MCL1 and mitofusin 1, two mitochondrial outer membrane proteins with short half-lives, have been identified as mitochondrial substrates of VCP-mediated MAD [23, 24]. In the HD cell cultures, we found that MCL1 is selectively degraded via the UPS pathway. Moreover, inhibition of VCP by either gene silencing or a dominant negative mutant recovered the MCL1 levels in HD mutant cells. These findings collectively suggest that mtHtt-induced MCL1 degradation depends upon VCP. VCP is required for Parkin-dependent CCCP-induced degradation of MFN1 and MFN2 and subsequent autophagy of dysfunctional mitochondria [22]. In our HD cell cultures, we found that the protein levels of MFN1 and MFN2 remained unchanged. Because of the lack of Parkin expression in HD mouse striatal cells [5], our findings may support the previous observation that the degradation of MFN1 and 2 requires the presence of Parkin [22, 23]. Thus, in the present study, MCL1 appears to be our best choice as a VCP mitochondrial substrate to define the signals of VCP-mediated MAD in the context of HD.
Under physiological conditions, VCP plays an important role in neurogenesis by promoting the formation of dendritic spines and synapses [45]. Mutations in the VCP gene are associated with several neurodegenerative diseases and causes mitochondrial dysfunction. Mice with homozygous pathogenic VCP mutations displayed mitochondrial abnormalities [46]. Fibroblasts derived from patients with VCP mutations exhibited mitochondrial depolarization and depletion of ATP content [47]. Additionally, over-expression of a disease-related VCP mutant in primary mouse neurons impaired clearance of damaged mitochondria [22]. Thus, VCP has been identified as an important factor in controlling mitochondrial quality. We recently found that mitochondria-accumulated VCP recruits the autophagic component LC3 to the mitochondria via the LC3-interacting regions (LIRs), which in turn triggers excessive mitophagy and mitochondrial degradation in various HD models [5]. Here, we further showed that VCP mitochondrial accumulation promotes MCL1 degradation in HD cultures. Depletion of MCL1 in the cortical neurons of mice predominantly activates an autophagic response prior to apoptotic induction [27]. Thus, it is likely that VCP has multiple molecular actions to promote mitochondria-associated autophagy. Whether and how these pathways crosstalk to regulate autophagy in HD remains to be determined.
Among VCP cofactors, UBXD1 seems to be unique in its interaction with VCP and is specifically affected by conditions associated with diseases [48, 49]. UBXD1 has been shown to locate VCP on endosomes, where it cooperates with VCP to regulate endolysosomal sorting of caveolin-1 [48]. In the present study, we found that UBXD1 was associated with mitochondria, where its interaction with VCP was enhanced in HD mutant cells. Moreover, UBXD1 selectively binds to MCL1 and the mitochondrial interaction between UBXD1 and VCP seems to be required to extract MCL1 from the mitochondria. Thus, UBXD1 might lend specificity to VCP-mediated MCL1 degradation in HD. Kim et al showed that UFD1L and NPL4 are recruited to the mitochondria in MEF and C2C12 cells exposed to CCCP and that VCP-UFD1L-NPL4 mediates MFN1 degradation [22]. Although we observed the translocation of UFD1L to the mitochondria in our HD mouse striatal cells, UFD1L did not bind to MCL1, suggesting that MCL1 is not the mitochondrial substrate for the VCP-UFD1L complex. The findings from our study and the previous report support the notion that VCP co-factors determine the specificity of VCP substrate recognition and degradation [19, 44]. VCP has about 40 identified cofactors in mammalian cells [50]; we cannot exclude the possibility that other co-factors participate in the VCP-mediated MCL1 degradation in HD.
Mitochondrial dysregulation is associated with neurodegeneration occurring during HD [51]. A key component of mitochondrial maintenance is quality control of damaged, dysfunctional proteins through protein turnover, which is likely the first line of defense mechanism against mitochondrial dysfunction in neurons [52, 53]. We found that the presence of mtHtt promotes mitochondrial protein degradation, thus it is conceivable that the failure in this defense mechanism in HD might be preferable to the destruction of whole mitochondrial organelle, which in turn lead to overall mitochondrial damage and neuronal cell death. This notion is support by our observation that the decrease in MCL1 protein level in HD cells resulted in mitochondrial depolarization, mitochondrial oxidative stress and enhanced autophagy, all of which are manifested in neuropathology of HD.
In summary, in this study we report an extensive degradation of the mitochondrial outer membrane protein MCL1 through a VCP-dependent pathway in the cell culture models of HD. We also identified UBXD1 as a VCP mitochondrial cofactor to facilitate MCL1 retro-translocation and degradation. The findings provide insights into the pathways of MAD, especially under disease conditions, and suggest an additional mechanism of mitochondrial dysfunction in HD.
Materials and Methods
Antibodies and reagents
Cycloheximide (CHX) and the protease inhibitor cocktail were purchased from Sigma–Aldrich. Proteasome inhibitor MG132 and VCP inhibitor DBeQ were from Tocris Bioscience. Antibodies against c-Myc (sc-40, 1:1000), GFP (sc-9996, 1:1000), HA (haemagglutinin, sc-7392, 1:1000), ubiquitin (sc-271289, 1:1000), HSP60 (sc-13115, 1:1000), MCL1 (sc-819, 1:1000), NPL4 (sc-134746, 1:1000), and p47 (sc-376614, 1:1000) were from Santa Cruz Biotechnology. VDAC (ab14734, 1:2000), VCP (ab109240, 1: 10,000), and UBXD1 (ab103651, 1:1000) antibodies were from Abcam. Anti-pan-actin antibody (A1978, 1: 10,000) was from Sigma–Aldrich. Anti-MFN1 (H00055669-M04, 1:1000) and MFN2 (H00009927-M01, 1:1000) antibodies were from Abnova and anti-UFD1L (NBP1-03339, 1:1000) was from Novus. Anti-mouse and Anti-rabbit IgG antibodies were from Thermo Scientific.
Constructs and transfection
Full-length VCP wild-type (VCPWT), VCP dominant negative mutant (VCPQQ), HA-UBXD1, GST-MCL1, and His-p47 were obtained from Addgene. Myc- or GFP-VCP was generated by inserting PCR-amplified fragments into the pCMV-Myc vector or GFP-N1 vector. Htt-Q23 and Q73 constructs were obtained from the CHDI Foundation. Cells were transfected with TransIT®-2020 (Mirus Bio LLC) following the manufacturer’s protocol.
Cell culture
Immortalized striatal cell lines HdhQ111 (mutant) and HdhQ7 (wild-type) derived from HdhQ111/111 and HdhQ7/Q7 knock-in transgenic mice (expressing 111 and 7 glutamine repeats, respectively) were obtained from the CHDI Foundation. Cells were cultured in DMEM supplemented with 10% fetal calf serum (FBS), 100µg/ml of penicillin, 100 µg/ml of streptomycin, and 400 µg/ml of G418. Cells were grown at 33°C in a 5% CO2 incubator. Cells with fewer than 14 passages were used in all experiments.
Human Embryonic Kidney HEK293T cells were maintained in DMEM supplemented with 10% FBS and 1% (v/v) penicillin/streptomycin.
HD patient fibroblasts (purchased from Coriell Institute, USA) and normal fibroblasts (purchased from Invitrogen) were maintained in MEM supplemented with 15% (vol/vol) heat-inactivated FBS and 1% (vol/vol) penicillin/streptomycin. All of the above cells were maintained at 37°C in 5% CO2–95% air.
Isolation of mitochondria-enriched fraction and lysate preparation
Cells were washed with ice-cold PBS and incubated on ice in a lysis buffer (250 mM sucrose, 20 mM Hepes/NaOH, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1:300 dilution protease inhibitor cocktail and 1:300 dilution phosphatase inhibitor cocktail) for 30 min. Cells were then scraped and disrupted by repeated aspiration through a 25-gauge needle. The homogenates were spun at 800 g for 10 min at 4°C, and the resulting supernatants were spun at 10,000 g for 20 min at 4°C. The pellets were washed with lysis buffer and spun again at 10,000 for 20 min at 4°C. The final pellets were suspended in lysis buffer containing 1% (v/v) Triton X-100 and were mitochondria-enriched fractions. Mitochondrial protein VDAC or HSP60 was used as a loading control.
RNA interference
Control siRNA, mouse VCP siRNA, and mouse UBXD1 siRNA were purchased from Thermo Fisher Scientific. HdhQ111 cells were transfected either with control siRNA or VCP siRNA or UBXD1 siRNA using TransIT-TKO® Transfection Reagent (Mirus Bio LLC), according to the manufacturer’s instructions.
Immunoprecipitation
Cells were lysed in a total cell lysate buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, and protease inhibitor) or in the mitochondrial isolation buffer described above. Total or mitochondrial lysates were incubated with the indicated antibodies overnight at 4°C followed by the addition of protein A/G beads for 1 hour. Immunoprecipitates were washed four times with cell lysate buffer and were analyzed by SDS-PAGE.
Mitochondrial membrane potential and ROS production
Cells cultured on coverslips were washed with PBS and then incubated with 0.25 µM tetra-methyl rhodamine (TMRM) (Invitrogen Life Science) for 20 minutes at 37°C. To ROS generation, cells were incubated with general ROS indicator (CM-H2DCFDA, Invitrogen Life Science) for 30 minutes at 37°C. The images were visualized by microscope and quantitation of the density of fluorescence was carried out using NIH ImageJ software. At least 100 cells per group were counted.
Western blot analysis
Protein concentrations were determined by Bradford assay. The protein was re-suspended in Laemmli buffer, loaded on SDS-PAGE, and transferred onto nitrocellulose membranes. Membranes were probed with the indicated antibodies, followed by visualization with ECL.
GST pull-down assay
Bacteria-expressed GST or GST–MCL1 were immobilized on glutathione-Sepharose 4B beads (GE Healthcare) for three hours and then washed three times. Beads were incubated with total lysates of mouse brains overnight at 4°C. Beads were then washed with a GST binding buffer (100 mM NaCl, 50 mM NaF, 2 mM EDTA, 1% Triton-X100, and protease inhibitor cocktail) and analyzed by SDS-PAGE.
Real-time PCR
Total RNA was isolated from the cells and reverse-transcribed with an oligo-dT primer. The following primers were used for real-time PCR: mouse MCL1 forward: 5’-AAAGGCGGCTGCATAAGTC-3’; mouse MCL1 reverse: 5’-TGGCGGTATAGGTCGTCCTC-3’; mouse GAPDH forward: 5’-TGGCCTTCCGTGTTCCTAC-3’; mouse GAPDH reverse: 5’-GAGTTGCTGTTGAAGTCGCA-3’.
Statistical analysis
Data were analyzed by Student’s t test or ANOVA with post-hoc Holm-Sidak test for comparison between two groups. Each study was performed with at least three independent replications. Data are expressed as mean ± SEM. Statistical significance was considered achieved when the value of p was < 0.05.
Highlights.
Mutant Htt promotes mitochondrial outer membrane protein MCL1 degradation via the UPS
Mutant Htt recruits VCP to the mitochondria, where VCP mediates MCL1 degradation
UBXD1 is a cofactor of VCP to facilitate MCL1 degradation in HD models
Acknowledgments
The work is supported by the National Institutes of Health grant (R01 NS088192) (to X.Q.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.The Huntington’s Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Guedes-Dias P, Pinho BR, Soares TR, de Proenca J, Duchen MR, Oliveira JM. Mitochondrial dynamics and quality control in Huntington’s disease. Neurobiol Dis. 2016;90:51–57. doi: 10.1016/j.nbd.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Reddy PH, Shirendeb UP. Mutant huntingtin, abnormal mitochondrial dynamics, defective axonal transport of mitochondria, and selective synaptic degeneration in Huntington’s disease. Biochim Biophys Acta. 2012;1822:101–110. doi: 10.1016/j.bbadis.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costa V, Scorrano L. Shaping the role of mitochondria in the pathogenesis of Huntington’s disease. EMBO J. 2012;31:1853–1864. doi: 10.1038/emboj.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo X, Sun X, Hu D, Wang YJ, Fujioka H, Vyas R, Chakrapani S, Joshi AU, Luo Y, Mochly-Rosen D, Qi X. VCP recruitment to mitochondria causes mitophagy impairment and neurodegeneration in models of Huntington’s disease. Nat Commun. 2016;7:12646. doi: 10.1038/ncomms12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo X, Disatnik MH, Monbureau M, Shamloo M, Mochly-Rosen D, Qi X. Inhibition of mitochondrial fragmentation diminishes Huntington’s disease-associated neurodegeneration. J Clin Invest. 2013;123:5371–5388. doi: 10.1172/JCI70911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bossy-Wetzel E, Petrilli A, Knott AB. Mutant huntingtin and mitochondrial dysfunction. Trends Neurosci. 2008;31:609–616. doi: 10.1016/j.tins.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damiano M, Galvan L, Deglon N, Brouillet E. Mitochondria in Huntington’s disease. Biochimica et biophysica acta. 2010;1802:52–61. doi: 10.1016/j.bbadis.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 9.Weydt P, Pineda VV, Torrence AE, Libby RT, Satterfield TF, Lazarowski ER, Gilbert ML, Morton GJ, Bammler TK, Strand AD, Cui L, Beyer RP, Easley CN, Smith AC, Krainc D, Luquet S, Sweet IR, Schwartz MW, La Spada AR. Thermoregulatory and metabolic defects in Huntington’s disease transgenic mice implicate PGC-1alpha in Huntington’s disease neurodegeneration. Cell Metab. 2006;4:349–362. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Panov AV, Gutekunst CA, Leavitt BR, Hayden MR, Burke JR, Strittmatter WJ, Greenamyre JT. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 11.Choo YS, Johnson GV, MacDonald M, Detloff PJ, Lesort M. Mutant huntingtin directly increases susceptibility of mitochondria to the calcium-induced permeability transition and cytochrome c release. Hum Mol Genet. 2004;13:1407–1420. doi: 10.1093/hmg/ddh162. [DOI] [PubMed] [Google Scholar]
- 12.Knott AB, Perkins G, Schwarzenbacher R, Bossy-Wetzel E. Mitochondrial fragmentation in neurodegeneration. Nat Rev Neurosci. 2008;9:505–518. doi: 10.1038/nrn2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hwang S, Disatnik MH, Mochly-Rosen D. Impaired GAPDH-induced mitophagy contributes to the pathology of Huntington’s disease. EMBO Mol Med. 2015;7:1307–1326. doi: 10.15252/emmm.201505256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 15.Taylor EB, Rutter J. Mitochondrial quality control by the ubiquitin-proteasome system. Biochem Soc Trans. 2011;39:1509–1513. doi: 10.1042/BST0391509. [DOI] [PubMed] [Google Scholar]
- 16.Karbowski M, Youle RJ. Regulating mitochondrial outer membrane proteins by ubiquitination and proteasomal degradation. Curr Opin Cell Biol. 2011;23:476–482. doi: 10.1016/j.ceb.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heo JM, Rutter J. Ubiquitin-dependent mitochondrial protein degradation. Int J Biochem Cell Biol. 2011;43:1422–1426. doi: 10.1016/j.biocel.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neutzner A, Youle RJ, Karbowski M. Outer mitochondrial membrane protein degradation by the proteasome. Novartis Found Symp. 2007;287:4–14. discussion 14–20. [PubMed] [Google Scholar]
- 19.Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol. 2012;14:117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- 20.Yamanaka K, Sasagawa Y, Ogura T. Recent advances in p97/VCP/Cdc48 cellular functions. Biochimica et biophysica acta. 2012;1823:130–137. doi: 10.1016/j.bbamcr.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 21.Heo JM, Livnat-Levanon N, Taylor EB, Jones KT, Dephoure N, Ring J, Xie J, Brodsky JL, Madeo F, Gygi SP, Ashrafi K, Glickman MH, Rutter J. A stress-responsive system for mitochondrial protein degradation. Mol Cell. 2010;40:465–480. doi: 10.1016/j.molcel.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim NC, Tresse E, Kolaitis RM, Molliex A, Thomas RE, Alami NH, Wang B, Joshi A, Smith RB, Ritson GP, Winborn BJ, Moore J, Lee JY, Yao TP, Pallanck L, Kundu M, Taylor JP. VCP is essential for mitochondrial quality control by PINK1/Parkin and this function is impaired by VCP mutations. Neuron. 2013;78:65–80. doi: 10.1016/j.neuron.2013.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu S, Peng G, Wang Y, Fang S, Karbowski M. The AAA-ATPase p97 is essential for outer mitochondrial membrane protein turnover. Mol Biol Cell. 2011;22:291–300. doi: 10.1091/mbc.E10-09-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perciavalle RM, Stewart DP, Koss B, Lynch J, Milasta S, Bathina M, Temirov J, Cleland MM, Pelletier S, Schuetz JD, Youle RJ, Green DR, Opferman JT. Anti-apoptotic MCL-1 localizes to the mitochondrial matrix and couples mitochondrial fusion to respiration. Nat Cell Biol. 2012;14:575–583. doi: 10.1038/ncb2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Bathina M, Lynch J, Koss B, Calabrese C, Frase S, Schuetz JD, Rehg JE, Opferman JT. Deletion of MCL-1 causes lethal cardiac failure and mitochondrial dysfunction. Genes Dev. 2013;27:1351–1364. doi: 10.1101/gad.215855.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Germain M, Nguyen AP, Le Grand JN, Arbour N, Vanderluit JL, Park DS, Opferman JT, Slack RS. MCL-1 is a stress sensor that regulates autophagy in a developmentally regulated manner. EMBO J. 2011;30:395–407. doi: 10.1038/emboj.2010.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas RL, Roberts DJ, Kubli DA, Lee Y, Quinsay MN, Owens JB, Fischer KM, Sussman MA, Miyamoto S, Gustafsson AB. Loss of MCL-1 leads to impaired autophagy and rapid development of heart failure. Genes Dev. 2013;27:1365–1377. doi: 10.1101/gad.215871.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas RL, Gustafsson AB. MCL1 is critical for mitochondrial function and autophagy in the heart. Autophagy. 2013;9:1902–1903. doi: 10.4161/auto.26168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okamoto T, Coultas L, Metcalf D, van Delft MF, Glaser SP, Takiguchi M, Strasser A, Bouillet P, Adams JM, Huang DC. Enhanced stability of Mcl1, a prosurvival Bcl2 relative, blunts stress-induced apoptosis, causes male sterility, and promotes tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 2013 doi: 10.1073/pnas.1321259110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mori M, Burgess DL, Gefrides LA, Foreman PJ, Opferman JT, Korsmeyer SJ, Cavalheiro EA, Naffah-Mazzacoratti MG, Noebels JL. Expression of apoptosis inhibitor protein Mcl1 linked to neuroprotection in CNS neurons. Cell death and differentiation. 2004;11:1223–1233. doi: 10.1038/sj.cdd.4401483. [DOI] [PubMed] [Google Scholar]
- 32.Trettel F, Rigamonti D, Hilditch-Maguire P, Wheeler VC, Sharp AH, Persichetti F, Cattaneo E, MacDonald ME. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet. 2000;9:2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- 33.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clausen TH, Lamark T, Isakson P, Finley K, Larsen KB, Brech A, Overvatn A, Stenmark H, Bjorkoy G, Simonsen A, Johansen T. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy. 2010;6:330–344. doi: 10.4161/auto.6.3.11226. [DOI] [PubMed] [Google Scholar]
- 35.Vicinanza M, Korolchuk VI, Ashkenazi A, Puri C, Menzies FM, Clarke JH, Rubinsztein DC. PI(5)P regulates autophagosome biogenesis. Mol Cell. 2015;57:219–234. doi: 10.1016/j.molcel.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ichimura Y, Kominami E, Tanaka K, Komatsu M. Selective turnover of p62/A170/SQSTM1 by autophagy. Autophagy. 2008;4:1063–1066. doi: 10.4161/auto.6826. [DOI] [PubMed] [Google Scholar]
- 37.Chou TF, Brown SJ, Minond D, Nordin BE, Li K, Jones AC, Chase P, Porubsky PR, Stoltz BM, Schoenen FJ, Patricelli MP, Hodder P, Rosen H, Deshaies RJ. Reversible inhibitor of p97, DBeQ, impairs both ubiquitin-dependent and autophagic protein clearance pathways. Proc Natl Acad Sci U S A. 2011;108:4834–4839. doi: 10.1073/pnas.1015312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 39.Schuberth C, Buchberger A. UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell Mol Life Sci. 2008;65:2360–2371. doi: 10.1007/s00018-008-8072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeung HO, Kloppsteck P, Niwa H, Isaacson RL, Matthews S, Zhang X, Freemont PS. Insights into adaptor binding to the AAA protein p97. Biochem Soc Trans. 2008;36:62–67. doi: 10.1042/BST0360062. [DOI] [PubMed] [Google Scholar]
- 41.Madsen L, Andersen KM, Prag S, Moos T, Semple CA, Seeger M, Hartmann-Petersen R. Ubxd1 is a novel co-factor of the human p97 ATPase. Int J Biochem Cell Biol. 2008;40:2927–2942. doi: 10.1016/j.biocel.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 42.Meyer HH, Shorter JG, Seemann J, Pappin D, Warren G. A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. The EMBO journal. 2000;19:2181–2192. doi: 10.1093/emboj/19.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kondo H, Rabouille C, Newman R, Levine TP, Pappin D, Freemont P, Warren G. p47 is a cofactor for p97-mediated membrane fusion. Nature. 1997;388:75–78. doi: 10.1038/40411. [DOI] [PubMed] [Google Scholar]
- 44.Buchberger A, Schindelin H, Hanzelmann P. Control of p97 function by cofactor binding. FEBS Lett. 2015;589:2578–2589. doi: 10.1016/j.febslet.2015.08.028. [DOI] [PubMed] [Google Scholar]
- 45.Wang HF, Shih YT, Chen CY, Chao HW, Lee MJ, Hsueh YP. Valosin-containing protein and neurofibromin interact to regulate dendritic spine density. J Clin Invest. 2011;121:4820–4837. doi: 10.1172/JCI45677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nalbandian A, Llewellyn KJ, Kitazawa M, Yin HZ, Badadani M, Khanlou N, Edwards R, Nguyen C, Mukherjee J, Mozaffar T, Watts G, Weiss J, Kimonis VE. The homozygote VCP(R(1)(5)(5)H/R(1)(5)(5)H) mouse model exhibits accelerated human VCP-associated disease pathology. PLoS One. 2012;7:e46308. doi: 10.1371/journal.pone.0046308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bartolome F, Wu HC, Burchell VS, Preza E, Wray S, Mahoney CJ, Fox NC, Calvo A, Canosa A, Moglia C, Mandrioli J, Chio A, Orrell RW, Houlden H, Hardy J, Abramov AY, Plun-Favreau H. Pathogenic VCP mutations induce mitochondrial uncoupling and reduced ATP levels. Neuron. 2013;78:57–64. doi: 10.1016/j.neuron.2013.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ritz D, Vuk M, Kirchner P, Bug M, Schutz S, Hayer A, Bremer S, Lusk C, Baloh RH, Lee H, Glatter T, Gstaiger M, Aebersold R, Weihl CC, Meyer H. Endolysosomal sorting of ubiquitylated caveolin-1 is regulated by VCP and UBXD1 and impaired by VCP disease mutations. Nat Cell Biol. 2011;13:1116–1123. doi: 10.1038/ncb2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stapf C, Cartwright E, Bycroft M, Hofmann K, Buchberger A. The general definition of the p97/valosin-containing protein (VCP)-interacting motif (VIM) delineates a new family of p97 cofactors. J Biol Chem. 2011;286:38670–38678. doi: 10.1074/jbc.M111.274472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meyer H, Weihl CC. The VCP/p97 system at a glance: connecting cellular function to disease pathogenesis. J Cell Sci. 2014;127:3877–3883. doi: 10.1242/jcs.093831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mochel F, Haller RG. Energy deficit in Huntington disease: why it matters. J Clin Invest. 2011;121:493–499. doi: 10.1172/JCI45691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neutzner A, Benard G, Youle RJ, Karbowski M. Role of the ubiquitin conjugation system in the maintenance of mitochondrial homeostasis. Ann N Y Acad Sci. 2008;1147:242–253. doi: 10.1196/annals.1427.012. [DOI] [PubMed] [Google Scholar]
- 53.Escobar-Henriques M, Langer T. Dynamic survey of mitochondria by ubiquitin. EMBO Rep. 2014;15:231–243. doi: 10.1002/embr.201338225. [DOI] [PMC free article] [PubMed] [Google Scholar]