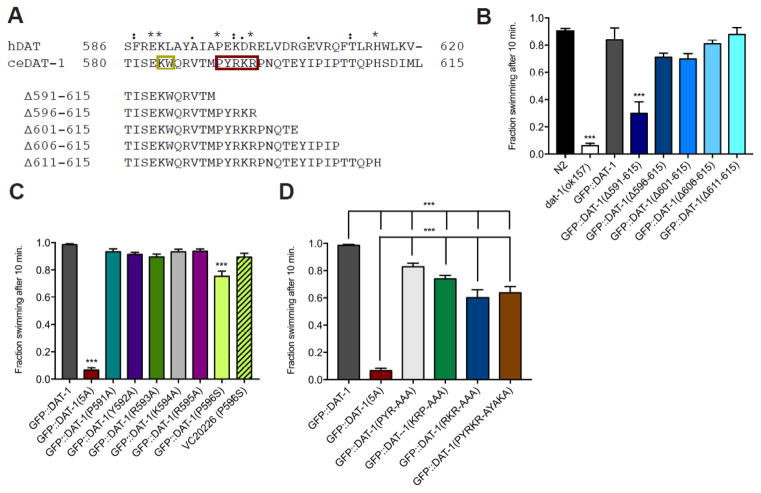
Figure 1. The 591PYRKR595 C-terminal motif is required for DAT-1 function.
A. Alignment of amino acids 580-615 of the DAT-1 C-terminus with sequences of the hDAT C-terminus. Identical (*), conservative (:) and semi-conservative (.) changes are noted. Amino acids retained in each deletion construct are indicated below the full sequence. Red box, novel 5 amino acid motif (PRYKR). Gold box, SEC-24 binding motif (KW).
B–D. Swip behavior of GFP::DAT-1 wild-type and mutations, narrowing down the C-terminal motif to 5 amino acids, PYRKR. All GFP::DAT-1 lines were produced on a dat-1(ok157) background, which is not noted. The figures represent the mean swimming behavior assessed by manual scoring, as described in Material and Methods. Data were analyzed using one-way ANOVA with multiple Bonferroni post-tests, comparing Swip levels of mutants to GFP::DAT-1 (B–D) or N2, (B) and with ***P<0.001 and error bars represent SEM.