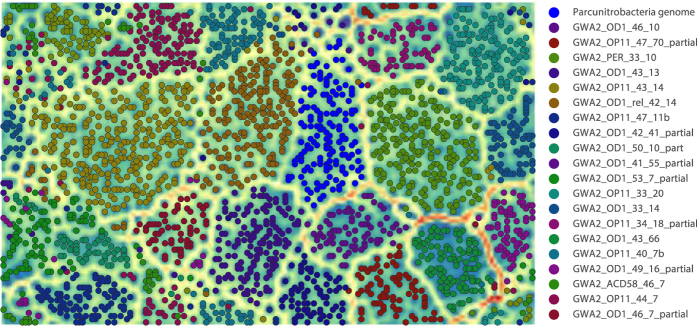
Figure 1. Validation of the Parcunitrobacteria draft genome using tetranucleotide frequencies and time-series abundance patterns.
Tetranucleotide frequencies and coverage were determined over 5 Kbp non-overlapping sliding windows for the Parcunitrobacteria genome along with a subset of genome bins from the GWA2 metagenome. The data were normalized and the ESOM was trained for 10 epochs using the Somoclu algorithm (https://arxiv.org/abs/1305.1422) using the option to initialize the codebook using PCA. Boundaries (dark bands) separate clusters of fragments with similar signatures (each dot represents a 5 kb fragment). The map was colored based on the binning information.