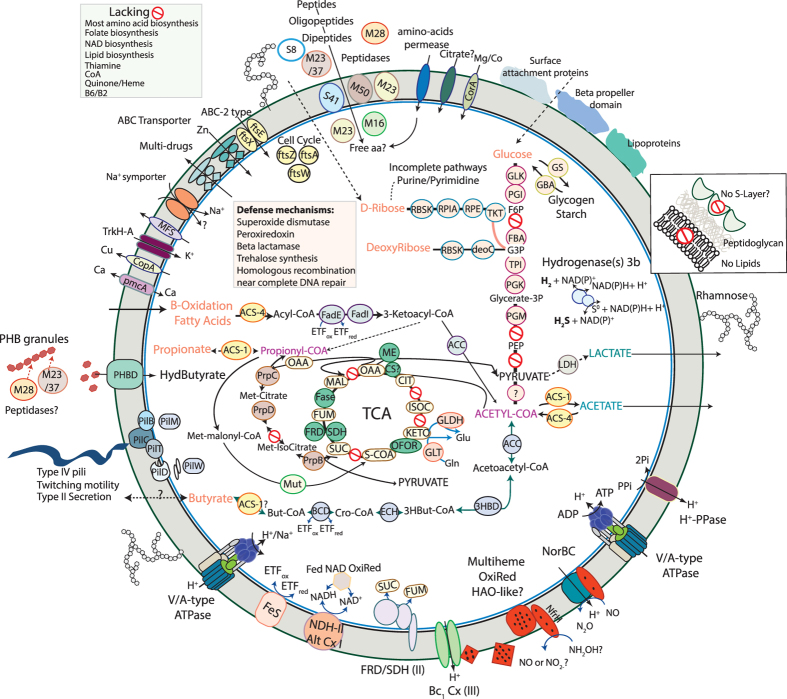
Figure 3. Energy metabolism of ‘Candidatus Parcunitrobacter nitroensis’.
Red “no entry” signs indicate genes missing from pathways. Abbreviations not defined in the text: TCA, tricarboxylic acid cycle; Glk, glucokinase; Pgi, glucose-6- phosphate isomerase; FBP, fructose-1,6-bisphosphatase; Pfk, phosphofructokinase; Aldo, fructose-bisphosphate aldolase; Pgk, phosphoglycerate kinase; TPI, triosephosphate isomerase; PHBD, polyhydroxybutyrate depolymerase; PGM, 1,3-bisphosphoglycerate-independent phosphoglycerate mutase; PPi, pyrophosphate; Pi, inorganic phosphate; OFOR, oxoglutarate/2-oxoacid-ferredoxin oxidoreductase; ACS, acetyl-CoA synthetase; LDH, lactate dehydrogenase; ME, malate dehydrogenase (oxaloacetate-decarboxylating); GS, glycogen synthase; GBA, glucosidase; RBKS, ribokinase; RpiA, ribose 5-phosphate isomerase A; RPE, ribulose-phosphate 3-epimerase; Tkt, transkelotase; deoC, deoxyribose-phosphate aldolase; GLDH, glutamate dehydrogenase; GLT, glutamate synthase (NADPH); Mut, methylmalonyl-CoA mutase;_rpC, 2-methylcitrate synthase; PrpD, 2-methylcitrate dehydratase; PrpB, methylisocitrate lyase; FadE, Acyl-CoA Dehydrogenase; FadJ, 3-hydroxyacyl-CoA dehydrogenase, enoyl-CoA hydratase; ACC, acetyl-CoA acetyltransferase or 3-ketoacyl-CoA thiolase; ETF, electron transfer flavoprotein; FeS, Electron-transferring-flavoprotein dehydrogenase/ubiquinone oxidoreductase; BCD, butyryl-CoA dehydrogenase; ECH, enoyl-CoA hydratase/isomerase; 3HBD, 3-hydroxybutyryl-CoA dehydrogenase; H+-PPase, proton-translocating pyrophosphatase; ATPase, ATP synthase; Cx I, NADH dehydrogenase; Cx II, succinate dehydrogenase/fumarate reductase; Fase, fumarate hydratase; CS, citrate synthase Cx III, bc1 complex; NorBC, nitric-oxide reductase; HOA, hydroxylamine oxidoreductase. The major classes of peptidases include: M50, M23, M16, M28, M23/37, S8 and S41 as presented in the figure. All of the protein sequences can be accessed at http://ggkbase.berkeley.edu/genome_summaries/1088-Parcunitrobacteria_curated_genome.