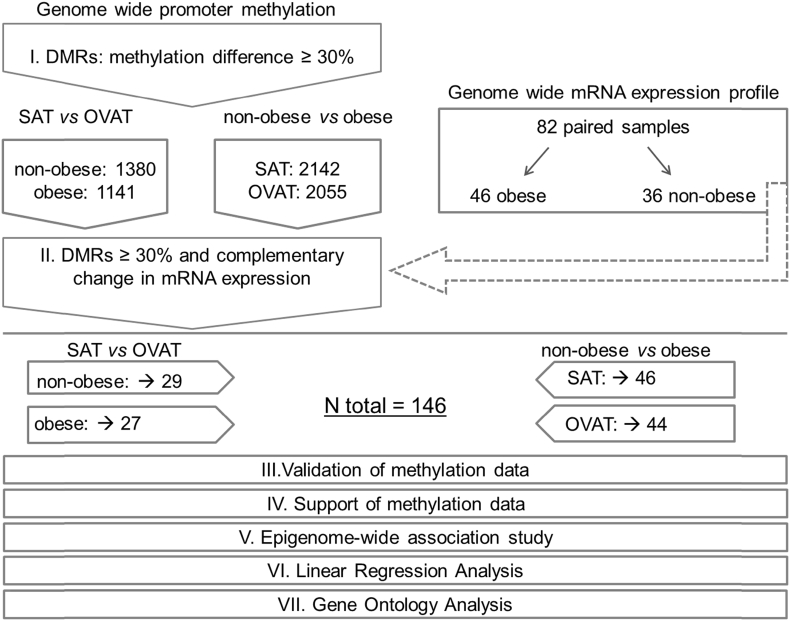
Figure 1.
Experimental workflow and study design. The figure shows the experimental workflow and study design. I. Genome wide promoter methylation was generated and an arbitrarily chosen cut off of 30% methylation differences (=DMR; differential methylated region) in the comparisons. Identified genes are given as numbers in non-obese and obese subgroups (SAT vs. OVAT) and in SAT and OVAT subgroups (non-obese vs. obese). II. The identified transcripts were tested for overlapping changes in mRNA expression. The final number of genes with negatively correlation between methylation and expression is underlined. Additionally, methylation data was validated using pyrosequencing for two genes (step III.) and supported in 3 independent data sets (step IV.). Furthermore, we conducted an epigenome-wide association study (EWAS) (step V.). Finally most promising candidates (N = 24) were tested for association with anthropometric and metabolic variables using linear regression analysis (step VI.) and analyzed using gene ontology analyses (step VII.).