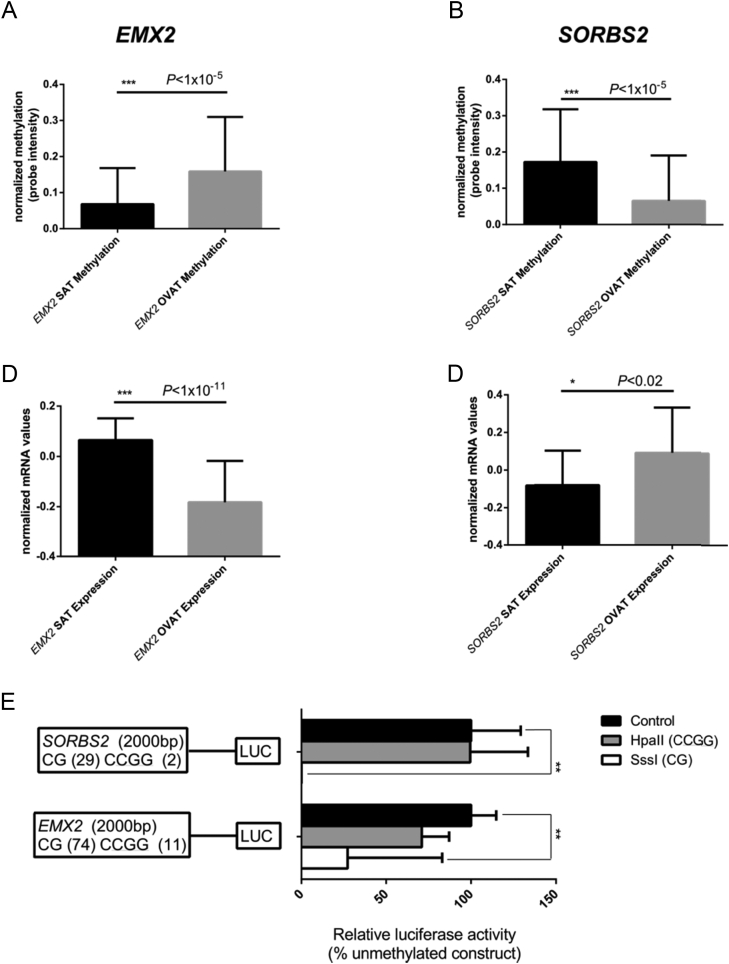
Figure 4.
Luciferase assays for SORBS2 and EMX2. The figure shows effect directions of methylation and expression in SAT and OVAT, respectively. Data shown represent results from the two luciferase reporter plasmids which were used to test effects of DNA methylation in SORBS2 and EMX2 promoter regions on transcriptional activity. Both plasmids comprised 2000 bp of either SORBS2 or EMX2 promoter regions inserted into a pCpGl-basic vector which was either methylated by HpaII (which methylates the internal Cs of the CCGG sequence) or SssI (which methylates all CpG sites) and then transfected into MCF7 cells. Data were normalized using a co-transfected renilla luciferase vector and presented as methylated promoter constructs (grey and white bars) relative to un-methylated constructs (black bars). Experiments were performed six times (SORBS) or eight times (EMX2) including four replicates in each experiment. Cells transfected with an empty pCpGL-basic vector and untransfected cells activities were used as background correction for firefly and renilla luciferase activity, respectively. *P < 0.05 **P < 0.005. Data are shown as mean ± SEM.