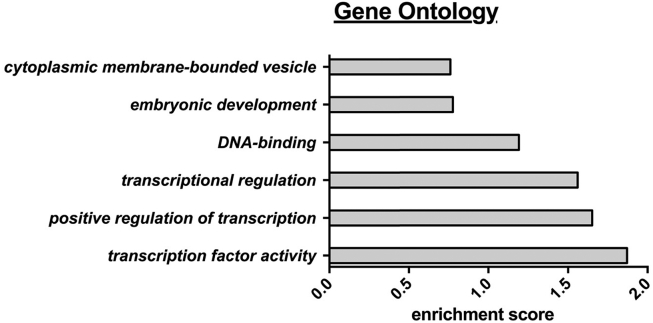
Figure 6.
Gene ontology analysis. Gene ontology analysis was performed using the DAVID program (http://david.abcc.ncifcrf.gov/summary.jsp) with default settings, high classification stringency and 0.5 as cut off for the enrichment score. Enriched pathways include the following genes: transcription factor activity: HAND2, PPARG, RUNX1, HSD17B8, HOXC6, TCF21; positive regulation of transcription: TCF21, MEOX1, HAND2, PPARG, RUNX1; transcriptional regulation: HOXC6, TCF21, MEOX1, HAND2, PPARG, ETV6, RUNX1, HSD17B8, AFF3; DNA-binding: HOXC6, TCF21, MEOX1, HAND2, PPARG, AFF3, ETV6, RUNX1.