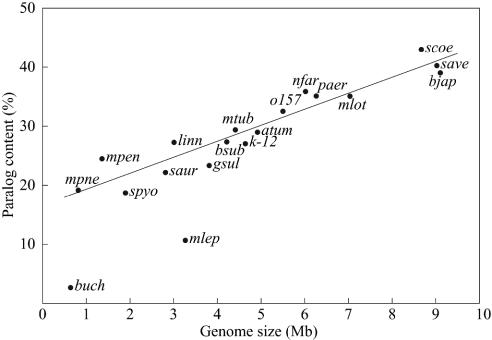
Fig. 3.
Correlation between paralog content and genome size in various bacteria: atum, Agrobacterium tumefaciens; bjap, Bradyrhizobium japonicum; bsub, Bacillus subtilis; buch, Buchnera sp.; o157, Escherichia coli O-157; k-12, E. coli K-12; gsul, Geobacter sulfurreducens; linn, Listeria innocua; mlot, Mesorhizobium loti; mtub, M. tuberculosis; mlep, Mycobacterium leprae; mpen, Mycoplasma penetrans; mpne, Mycoplasma pneumoniae; nfar, N. farcinica; paer, Pseudomonas aeruginosa; saur, Staphylococcus aureus; spyo, Streptococcus pyogenes; save, S. avermitilis; and scoe, S. coelicolor A3 (2). A linear regression line, which was estimated without using buch and mlep data, is also indicated. Extremely low paralogies in buch and mlep are due to massive decay of genes in these genomes.