Abstract
Hepatitis B virus (HBV) e (HBe) antigen is a nonstructural virus component with great immune regulation roles. It regulates adaptive immunity response and participates in persistent infection development. However, its roles on monocytes and B lymphocytes were rarely studied. Herein, we studied HBe roles on U937 and Hmy2.CIR by creating HBe stably transfected cells using lentivirus. We detected the motility of HBe-U937 through transwell migration assay. Cytokines that primarily produced by monocytes, including BAFF, B-cell activating factor (BAFF), interleukin (IL)-6, IL-10, tumor necrosis factor (TNF)-α, and A proliferation inducing ligand (APRIL), were measured in culture supernatants of transfected U937, and serum BAFF, IL-6, and IL-10 were detected in HBe-positive and HBe-negative HBV-infected patients. Among these, BAFF mRNA and membrane-bound BAFF were further detected. Activation and inhibition markers of B lymphocytes on HBe-Hmy2.CIR and proliferation of transfected Hmy2.CIR after coculture with transfected U937 were also detected. We found that U937 migration was inhibited by HBe. BAFF expression was increased in HBe-U937, however, membrane-bound BAFF on HBe-U937 was decreased. In addition, Serum BAFF in HBe-positive patients was higher than in HBe-negative patients. IL-6 and IL-10 were increased in HBe-U937 after being stimulated by lipopolysaccharide (LPS), however, serum IL-6 and IL-10 were not associated with HBe status in patients. Besides, TNF-α and APRIL expression were basically the same in GV166-U937 and HBe-U937. B lymphocyte activation markers CD86 and Tspan33 were raised in HBe-Hmy2.CIR. However, inhibition markers Lyn and CD32b had no differences between HBe-Hmy2.CIR and control. Proliferation of transfected Hmy2.CIR was not affected by coculture with transfected U937, however, HBe transfection itself enhanced Hmy2.CIR proliferation. Altogether, these revealed that HBe can inhibit U937 migration and promote cytokines, including BAFF, IL-6, and IL-10, production in U937. Besides, HBe enhances BAFF release from U937 and increases BAFF concentration in vivo. In addition, HBe antigen facilitates Hmy2.CIR activation and proliferation through direct induction.
Keywords: : HBV e antigen, monocyte, B lymphocyte, cytokine, B cell activating factor
Introduction
Hepatitis B virus (HBV) is the main cause of acute or chronic liver diseases; chronic HBV infection can develop into liver cirrhosis with the potential sequela of hepatocellular carcinoma (HCC) (8). Currently, although cellular adaptive immunity in HBV infection is well reported, the innate immunity response to HBV is still not clear and is difficult to study (1). HBV is not strictly hepatotropic, and except for hepatocytes, it infects peripheral blood mononuclear cells (PBMCs). It has been reported that PBMCs have higher HBV infection rates in monocytes and B lymphocytes than in T lymphocytes and NK cells (27,31). However, little information is available at present about influences of HBV on the function of monocyte and B lymphocyte during HBV infections (2,3,12).
HBV e (HBe) antigen, also known as pre-Core protein, is a nonstructural, secretory protein encoded by HBV Core open reading frames (30). It is a remarkable immune regulator in adaptive and innate immune response in the host against HBV processes. During the innate immune response, toll-like receptor 2 (TLR2) expression levels on monocytes from HBe-positive patients were significantly decreased compared with HBe-negative patients, and cytokines like tumor necrosis factor (TNF) and interleukin (IL)-6 produced by monocytes upon stimulation with TLR2 agonists were reduced in HBe-positive patients in vitro, which indicated that HBe can inhibit monocyte function (33). Recently, researchers have demonstrated that recombinant HBe inhibits the mobility of human monocytes in vitro, also prompting a repressive role for HBe in monocyte function (20). Despite all that, impacts of HBe on monocytes and B lymphocytes still need more exploration.
Although the capability of HBV to replicate in extrahepatic cells remains controversial, its DNA is truly detectable in these cells. To simulate natural processes of HBe in monocytes and B lymphocytes, and further study roles of HBe on these cells, we transfected U937 (monocyte cell line) and Hmy2.CIR (B lymphocyte cell line) with lentiviral vectors harboring HBe gene. Then, the motility and the B cell-activating factor (BAFF) expression level of stably transfected U937 cells were detected, because monocyte migration is critical in innate immune initiation and BAFF is an important B lymphocyte-activating factor, which is primarily secreted by monocytes. Other cytokines that are primarily produced by monocytes, including proinflammatory factors TNF-α, IL-6, anti-inflammatory factor IL-10, and another B lymphocyte stimulator A proliferation inducing ligand (APRIL), were also detected. In addition, the activation status of stably transfected Hmy2.CIR cells was measured, and proliferation of transfected Hmy2.CIR in coculture systems with HBe-positive or HBe-negative U937 was determined.
Materials and Methods
Reagents
All standard culture reagents were obtained from Gibco (Life Technologies), Carlsbad, CA. Puromycin was purchased from Sigma-Aldrich (Shanghai) Trading Co., LTD. FITC-labeled monoclonal antibodies specific to BAFF and B-cell activating factor receptor (BAFF-R) were purchased from eBioscience, USA. Monoclonal antibodies specific to HBe was purchased from Abcam (Hong Kong) Ltd., New Territories, Hong Kong. The reverse transcription polymerase chain reaction (PCR) kit and SYBR green master used for semiquantitative real time-PCR were purchased from Roche Diagnostics Corporation, Roche Applied Science, Indianapolis, IN. BAFF, IL-6, and TNF-α enzyme-linked immunosorbent assay (ELISA) kits were purchased from R&D Systems, Inc., Minneapolis, MN. IL-10 and APRIL ELISA kits were purchased from Abcam. 6.5 mm Transwell® with 0.4 μm pore polycarbonate membrane inserts was purchased from Corning. Cell Counting Kit-8 was from Dojindo Laboratorise (Shanghai), Co., Ltd. All primers were synthesized and validated by Sangon Biotech (Shanghai, China) Co., Ltd.
Cell culture and transfection
U937 and Hmy2.CIR cells were obtained from Shanghai Institute for Biological Sciences, Chinese Academy of Sciences. U937 cells were grown in the RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), penicillin (100 IU/mL), and streptomycin (100 IU/mL). Hmy2.CIR cells were grown in the IMDM medium supplemented with 10% FBS and antibiotics. Cells were cultured in a humidified incubator with 5% CO2 at 37°C.
HBe lentiviral vectors were constructed and packaged by Shanghai Genetimes Technology, Inc.; these lentiviruses were puromycin resistant. U937 cells and Hmy2.CIR cells (1 × 104) were seeded in 96-well tissue culture plates and infected with lentiviral vectors at multiplicities of infection (MOI) of 20, with the empty lentiviral vector GV166 as the negative control. Then, cells were infected for 8 h. Subsequently, the medium was replaced with a fresh complete medium. After 2 days, these transfected U937 cells and Hmy2.CIR cells were screened with 2 μg/mL puromycin in 1640 and IMDM medium supplemented with 10% FBS, respectively. Five days later, the sorted living cells were subcultured and used for experiments.
Identification of HBe DNA and mRNA in transfected cells
The transfected cells were sorted and harvested for DNA and RNA isolation, respectively. Genome DNA was extracted from cells with a standard salting-out procedure. Total RNA was isolated from cells by using Tripure isolation reagent (Roche, USA). Genome DNA and total RNA concentration were determined using ultraviolet spectrophotometer DU800 (Beckman), and RNA concentration was normalized to 200 ng/μL. Then, total RNA was converted into cDNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche, USA). PCR primers used for HBe DNA amplification were as follows: HBe sense: 5′-ATGGACATTGACCCGTATAAAG-3′; HBe anti-sense: 5′-CTAACATTGAGATTCCCGAGATTG-3′. Reverse transcription PCR primers used for HBe cDNA amplification were as follows: HBe sense: 5′-CTA TTC TGT GTT GGG GTG AG-3′; HBe anti-sense: 5′-AAG TAA GGC AGG AAA TGT GA-3′. In detail, 50 ng genome DNA or 1 μL cDNA was amplified as template in 20 μL PCR system containing 1 mM dNTP, 0.2 mM MgCl2, 500 nM primer, and 0.5 U Taq polymerase. Cycling was carried out in a 9700 thermal cycler (PE Applied Bio systems) under the following conditions: 5 min at 95°C, 33 cycles of 95°C for 30 sec, 60°C for 30 sec, and 72°C for 45 sec, followed by a final extension step for 7 min at 72°C. The PCR system without DNA was set as negative control. PCR products were analyzed by 1.5% ethidium bromide–agarose gel electrophoresis.
Immunofluorescence assay
To detect whether HBe protein was expressed in transfected Hmy2.CIR cells, we performed immunofluorescence on transfected Hmy2.CIR cell climbing pieces. Cell climbing pieces were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min, permeabilized with 0.5% Triton X-100 in PBS for 2 min, followed by 30 min incubation with goat serum. Then, cell climbing pieces were washed and incubated with rabbit anti-HBe primary antibodies overnight at 4°C. After being washed by PBS, cell climbing pieces were incubated with FITC-labeled goat anti-rabbit IgG for 1 h at room temperature and finally stained with DAPI. The fluorescence signals were visualized by fluorescence microscopy (Nicon Eclipse Ti-S, Japan) and analyzed using Nis-Elements D 3.2 software.
HBe antigen assessment by Elecsys
HBe-U937 and HBe-Hmy2.CIR cells were collected and lysed in the Pierce RIPA buffer (Thermo, USA). Total proteins were extracted from cell lysate after homogenization and centrifugation. The protein concentration was determined using the BCA protein assay kit (Pierce). At the same time, cell culture supernatants were collected. Then, the extracted total proteins and culture supernatants were used for HBe antigen detection through electrochemical luminescence assay according to the manual operation.
Cell migration experiment
An aliquot of 5 × 105 cells (U937, GV166-U937, and HBe-U937 cells) were resuspended in serum-free RPMI 1640 medium and seeded at the upper chamber of 8 μm polycarbonate transwell inserts (Corning, Inc., Lowell, MA) in 24-well plates. The bottom chamber contained RPMI 1640 with 10% FBS. After 7 h, cells migrated into the lower chamber were collected and counted. The migration percentage was calculated as the numbers of cells migrating into the bottom chamber added to the number of cells adhering to the polycarbonate and then divided by the number of cells collected from a well that is lacking an insert.
Semiquantitative RT-PCR
BAFF expression levels in U937- and U937-transfected cells (GV166-U937, HBe-U937), B lymphocyte activation markers CD69, CD86, and TSPAN33, and inhibition molecules Lyn, CD32b expression levels in Hym2.CIR and transfected cells (GV166-Hmy2.CIR, HBe-Hmy2.CIR) with or without CD40L (1.5 μg/mL), and IL-4 (3 ng/mL) stimulation were measured by semiquantitative RT-PCR. Total RNA extraction and cDNA reverse transcription were performed as mentioned before, and RT-PCR was performed in 20 μL solution with the gene-specific primers described as follows:
BAFF sense: 5′-ATGTATTCAAAATATGCCTGA-3′;
BAFF anti-sense: 5′-ATGTGACATCTCCATCCAGTG-3′;
CD69 sense: 5′-AGAACAGCTCTTTGCATCCG-3′;
CD69 anti-sense: 5′-TGGTGATGAAGACCACATTCA-3′;
CD86 sense: 5′-ATGAGTGGGGTCATTTCCAG-3′;
CD86 anti-sense: 5′-AGAGGAGCAGCACCAGAGAG-3′;
TSPAN33 sense: 5′-ATGTTCCTGCTCACCTTCTG-3′;
TSPAN33 anti-sense: 5′-AGCCTTGTCTGAGAAGACGA-3′;
Lyn sense: 5′-GCCCATTTACATCATCACCG-3′;
Lyn anti-sense: 5′-AGCACTTTGCCACCTTCATC-3′;
CD32b sense: 5′-GCGGCCATTGTTGCTGCTGT-3′;
CD32b anti-sense: 5′-AGAGCATCCGGGTGCATGAGA-3′.
GAPDH was PCR amplified for normalization in all reactions using the following primers:
GAPDH sense: 5′-AACAGCCTCAAGATCATCAGC-3′;
GAPDH anti-sense: 5′-GGATGATGTTCTGGAGAGCC-3′.
Enzyme-linked immunosorbent assay
An aliquot of 1 × 106 U937- and U937-transfected cells (GV166-U937, HBe-U937) were seeded in six-well plates. After a 24-h culture, the culture supernatant was collected and BAFF in culture supernatant were analyzed by a commercially available Quantikine Human BAFF/BLyS ELISA kit (R&D Systems, Inc.), according to the manufacturer's instructions. For IL-6, IL-10, TNF-α, and APRIL detection, U937- and U937-transfected cells (GV166-U937, HBe-U937) were seeded in six-well plates as mentioned above, and induced with lipopolysaccharide (LPS) (1 μg/mL) for 24 h before culture supernatant collection. IL-6, IL-10, TNF-α, and APRIL in these culture supernatants were analyzed by commercially available ELISA kits, including Quantikine ELISA Human IL-6 kit (R&D Systems, Inc.), Quantikine ELISA Human TNF-α kit (R&D Systems, Inc.), IL-10 Human SimpleStep ELISA kit (Abcam), and APRIL Human ELISA kit (Abcam). In addition, serum BAFF, IL-6, and IL-10 were measured in samples from 16 HBe-positive and 15 HBe-negative newly diagnosed patients with hepatitis B in our hospital by using the same ELISA kit.
Flow cytometric analysis
To assess cell surface molecules, flow cytometry was employed to analyze membrane-bound BAFF on surface of U937-transfected cells and BAFF-R on surface of Hmy2.CIR-transfected cells. An aliquot of 1 × 106 cells was prepared and the surfaces were stained as per standard protocol. Stained cells were run on flow cytometry machine (Epics XL-4; Beckman Coulter). Briefly, cells were collected, washed twice with PBS, and 200 μL of cells was dispensed into two tubes for each sample, that is, one assay tube and one control tube. Then, 20 μL of conjugated specific antibodies was added to each assay tube (FITC-conjugated specific anti-BAFF antibody and anti-BAFF-R antibody; eBiosience) and corresponding FITC-conjugated isotype control antibodies were added to control tubes. Tubes were incubated for 15 min at room temperature (18–25°C) in the dark. Subsequently, 3 mL PBS was added into each tube, centrifuged for 5 min at 300 g at room temperature, supernatant was discarded, and cells were resuspended in 500 μL PBS and subjected to flow cytometry analysis.
Cell proliferation
An aliquot of 5 × 104 HBe-positive (HBe-Hmy2.CIR) or HBe-negative (GV166-Hmy2.CIR) Hmy2.CIR cells were seeded in down chambers of the 24-well coculture systems with 600 μL media, and 5 × 104 HBe-positive (GV166-U937) or HBe-negative (HBe-U937) U937 cells were seeded in up inserts with 0.4 μm pore polycarbonate membrane with 100 μL media. These cells were cocultured for indicated times in IMDM media with 10% FBS, penicillin (100 IU/mL), and streptomycin (100 IU/mL). After that, inserts were removed and proliferation of HBe-positive or HBe-negative Hmy2.CIR cells was detected by using Cell Counting Kit-8 (Dojindo Laboratorise, co., LTD).
Statistical analysis
All experiments were performed in triplicate. Data from multiple experiments were averaged and expressed as mean ± SD. Statistical comparisons were performed using ANOVA and Dunnett's t-test. Dunnett's t-test was used to determine whether there is a significant difference between HBe-transfected group and control group, when the p-value of ANOVA indicated that at least two groups differ from each other in our study. Statistical significance was set at a p-value <0.05.
Results
HBe DNA fragment was successfully integrated into and expressed in U937 and Hmy2.CIR cells
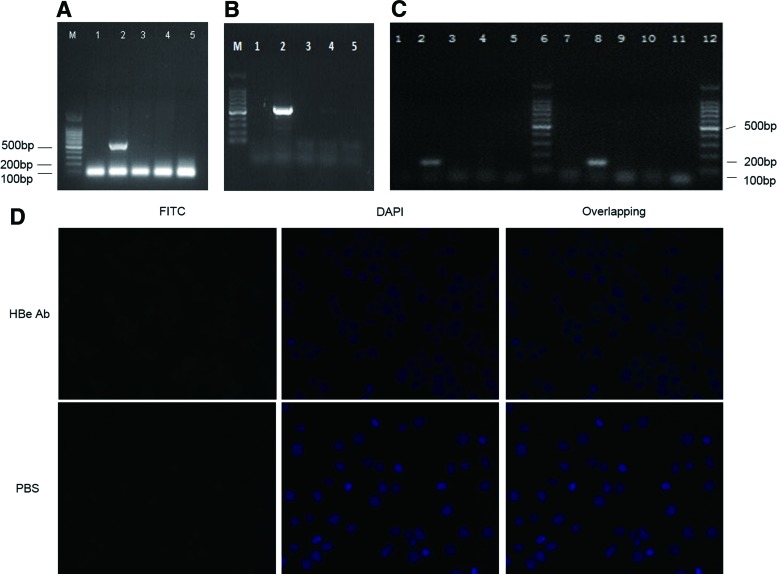
Agarose gel electrophoresis of PCR product showed that the HBe gene was successfully integrated into U937 and Hmy2.CIR cellular genomes, respectively (Fig. 1A, B). Reverse transcription PCR confirmed that HBe mRNA was transcribed in transfected U937 and Hmy2.CIR cells, respectively (Fig. 1C). Through immunofluorescence staining, we proved that HBe expressed weekly in cytoplasm of HBe-Hmy2.CIR cells, and almost more than 90% cells were positive for HBe expression (Fig. 1D). Elecsys demonstrated that the cutoff index (COI) of cultured supernatant of HBe-U937 and HBe-Hmy2.CIR was 3.00 and 4.00, respectively, and the COI of total protein extracted from HBe-U937 and HBe-Hmy2.CIR was 33.00 and 34.00, respectively. This result indicated that HBe antigen was detectable both in HBe-U937 and HBe-Hmy2.CIR cells, further confirming the result of immunofluorescence staining.
FIG. 1.
HBe was successfully transfected into U937 and Hmy2.CIR cells and expressed in these cells. (A) DNA extracted from Hmy2.CIR and transfected Hmy2.CIR cells were used as templates to amplify HBe DNA fragments. The result of DNA agarose gel electrophoresis showed that HBe DNA fragment was successfully integrated in Hmy2.CIR cells. Lane 1: empty lentivirus carrier—Hmy2.CIR; Lane 2: HBe-Hmy2.CIR; Lane 3: Hmy2.CIR; Lane 4 and Lane 5: negative control. (B) DNA extracted from U937 and transfected U937 cells were used as templates to amplify HBe DNA fragments. DNA agarose gel electrophoresis showed that HBe DNA fragment was successfully integrated in U937 cells also. Lane 1: empty lentivirus carrier—U937; Lane 2: HBe-U937; Lane 3: U937; Lane 4 and Lane 5: negative control. HBe DNA fragment is 552 bp in length. (C) HBe mRNA expression in Hmy2.CIR and U937 cells detected by reverse transcription PCR. Lane 1: empty lentivirus carrier—Hmy2.CIR; Lane 2: HBe-Hmy2.CIR; Lane 3: Hmy2.CIR; Lane 4 and Lane 5: negative control; Lane 6: DNA Marker; Lane 7: empty lentivirus carrier—U937; Lane 8: HBe-U937, Lane 9: U937; Lane 10 and Lane 11: negative control; Lane 12: DNA marker. The product of the reverse transcription PCR is 156 bp in length. This result proved that HBe DNA was successfully transcribed in transfected Hmy2.CIR and U937 cells. (D) Expression of HBe protein in transfected Hmy2.CIR cells detected by immunofluorescence. There was weak fluorescence in cytoplasm of HBe-Hmy2.CIR cells after incubation with HBe antibody and FITC-labeled second antibody (the first lane). It verified that HBe protein was weakly expressed in cytoplasm of HBe-Hmy2.CIR cells. Hmy2.CIR cells incubated with PBS was done as negative control. PCR, polymerase chain reaction. Color images available online at www.liebertpub.com/vim
HBe inhibited U937 migration
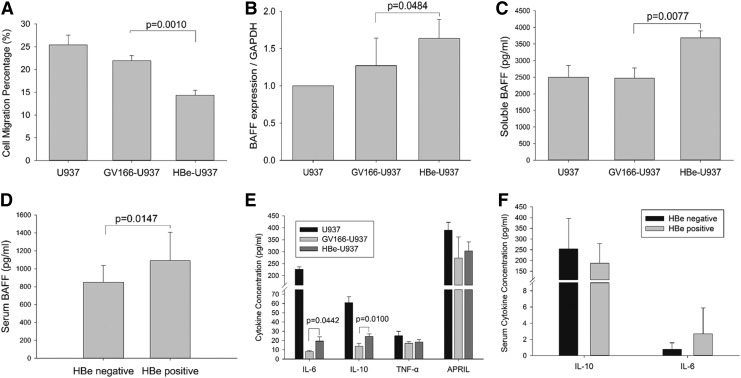
Macrophages, from the monocyte differentiation, play an important role in innate immune response, defending virus infection. To understand the ability to enrich monocytes into tissues and organs, we further investigated the migration of U937 cells using transwell inserts. The result revealed that HBe-U937 cells displayed significantly decreased migration rates compared with U937 cells (GV166-U937 vs. HBe-U937: 21.9% vs. 14.4%; p = 0.0010; Fig. 2A), which indicated an inhibitory role of HBe in U937 motility.
FIG. 2.
HBe inhibited U937 motility and increased BAFF, IL-6, and IL-10 expression in monocytes. (A) Impaired migration ability of HBe-transfected U937 cells. The percentage of migratory cells in HBe-U937 group was sufficiently decreased when compared with GV166-HBe group (GV166-U937 vs. HBe-U937: 21.9% vs. 14.4%; p = 0.0010). This revealed a significant inhibitory role of HBe in the migration ability of U937 cells. (B) BAFF mRNA expression in HBe-transfected U937 cells. Semiquantitative RT-PCR showed that the BAFF mRNA expression level in HBe-U937 cells was significantly increased, which indicated that HBe promoted BAFF expression in U937 cells. GAPDH was used for reference gene to normalize BAFF mRNA level. (C) Soluble BAFF concentration in culture supernatants of U937 and transfected U937 cells. It showed that soluble BAFF secretion was increased in HBe-transfected U937 cells. (D) Serum BAFF levels in HBe-positive and HBe-negative patients with HBV infection. Serum samples were collected from patients and BAFF levels were measured by ELISA. This result confirmed that BAFF secretion increased in HBe-positive patients. (E) Other cytokines in culture supernatant of U937 and transfected U937 cells. IL-6 and IL-10 expression in lentivirus-transfected U937 were inhibited compared with U937. However, when compared with GV166-U937, it showed that both IL-6 and IL-10 were increased in HBe-U937 cells. TNF-α and APRIL expression had no significant difference between groups. (F) Serum IL-6 and IL-10 in HBe-positive and HBe-negative CHB patients. It showed that IL-6 and IL-10 expression were not associated with HBe antigen status in HBV-infected patients. All experiments were repeated thrice independently. Differences between groups were analyzed using the independent sample Student's t-test and considered to be significant if p < 0.05. APRIL, A proliferation inducing ligand; BAFF, B cell activating factor; CHB, chronic hepatitis B; ELISA, enzyme-linked immunosorbent assay; IL, interleukin; RT, real time; TNF, tumor necrosis factor.
HBe enhanced BAFF expression in U937 cells
BAFF is a critical B lymphocyte activation factor, which is produced mainly by monocytes (26). We detected BAFF expression in transfected U937 cells. Results of semiquantitative RT-PCR displayed that mRNA level of BAFF in the HBe-U937 group was increased significantly compared with GV166-U937 control (HBe-U937 vs. GV166-U937, p = 0.0484; Fig. 2B). In addition, we measured the concentration of soluble BAFF in culture supernatant of U937 and transfected U937 cells. As shown in Figure 2, soluble BAFF in culture supernatant of HBe-U937 were elevated significantly compared to the GV166-U937 control group (HBe-U937 vs. GV166-U937: 3903.33 ± 205.51 pg/mL vs. 2710.00 ± 310.00 pg/mL, p = 0.0077; Fig. 2C). These results showed that BAFF expression in U937 cells was elevated by HBe. Furthermore, serum BAFF measured in HBe-positive and HBe-negative patients with hepatitis B demonstrated that the average BAFF level in HBe-positive patients was significantly higher than in HBe-negative patients (1092.28 ± 315.08 pg/mL vs. 848.33 ± 188.19 pg/mL, p = 0.0147; Fig. 3D), which confirmed that HBe can increase BAFF in vivo.
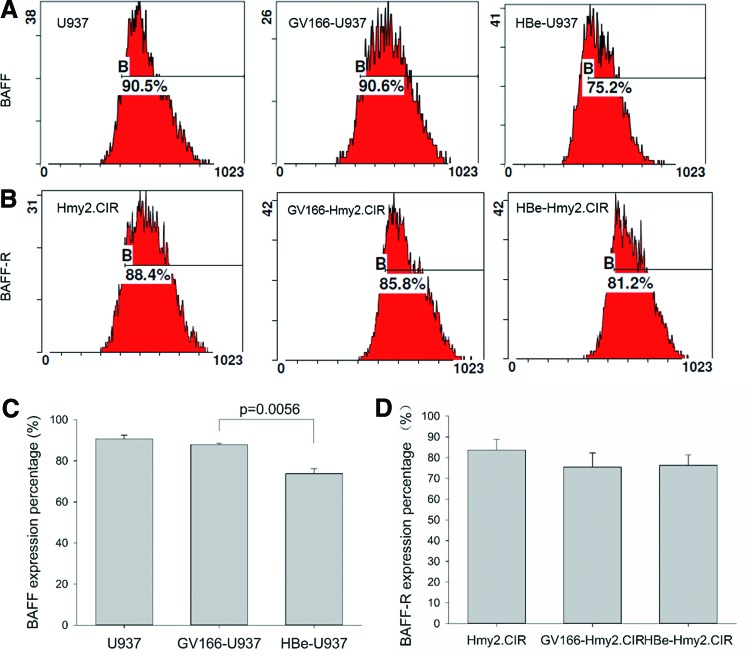
FIG. 3.
Membrane-bound BAFF on U937 cells and BAFF-R on Hmy2.CIR cells detected by flow cytometry. (A) Presentation of one single experiment about mBAFF expression on U937 and transfected U937 cells detected by flow cytometry. (B) Presentation of one single experiment about BAFF-R expression on Hmy2.CIR and transfected Hmy2.CIR cells detected by flow cytometry. (C) mBAFF expression on U937 and transfected U937 cells. Statistical results of triple experiments about mBAFF expression on U937 and transfected U937 cells were presented, which revealed that there was a significant decrease in mBAFF expression on HBe-U937 cells (GV166-U937 vs. HBe-U937: 87.83% vs. 73.77%, p = 0.0056). (D) Statistical results of triple experiments about BAFF-R expression on Hmy2.CIR and transfected Hmy2.CIR cells. There was no significant difference in BAFF-R expression on HBe-Hmy2.CIR cells compared with Hmy2.CIR cells. All experiments were repeated thrice independently. Differences between groups were analyzed using the independent sample Student's t-test and considered to be significant if p < 0.05. BAFF-R, B-cell activating factor receptor; mBAFF, membrane-bound BAFF. Color images available online at www.liebertpub.com/vim
HBe promoted BAFF releasing from membrane of U937 cells
We measured membrane-bound BAFF on U937 and transfected U937 cells by flow cytometry, which revealed that the membrane bonding BAFF significantly decreased in the HBe-U937 group compared with that in GV166-U937 control group (GV166-U937 vs. HBe-U937: 87.83% vs. 73.77%, p = 0.0056; Fig. 3A, C). Since BAFF mRNA and soluble BAFF were increased in HBe-U937 cells, this result indicated a possible role of HBe in regulating the kinetics of BAFF release that HBe promoted BAFF release from U937 cells.
Meanwhile, BAFF-R on membranes of HBe-Hmy2.CIR cells was measured; there was no significant difference compared with GV166-Hmy2.CIR cells (Fig. 3B, D).
HBe enhanced IL-6 and IL-10 production in U937 with LPS induction
We further detected other cytokines primarily produced by monocytes, including proinflammatory factors TNF-α and IL-6, anti-inflammatory factor IL-10, and another B lymphocyte stimulator APRIL in U937, and HBe-transfected U937 cells, after being stimulated by LPS. Results proved that IL-6 and IL-10 expression were significantly inhibited by lentivirus transfection (Fig. 2E). However, when compared with GV166-U937, IL-6 and IL-10 expression significantly increased in HBe-U937 (IL-6: GV166-U937 vs. HBe-U937: 8.05 ± 1.25 pg/mL vs. 19.39 ± 4.67 pg/mL, p = 0.0442; IL-10: GV166-U937 vs. HBe-U937: 13.79 ± 3.06 vs. 24.57 ± 2.47 pg/mL, p = 0.0100; Fig. 2E). TNF-α and APRIL expression were basically the same between GV166-U937 and HBe-U937 groups (TNF-α: GV166-U937 vs. HBe-U937: 17.06 ± 1.70 pg/mL vs. 18.27 ± 2.70 pg/mL, p = 0.5525; APRIL: GV166-U937 vs. HBe-U937: 273.63 ± 87.73 vs. 302.74 ± 38.76 pg/mL, p = 0.6384; Fig. 2E). Serum IL-6 and IL-10 concentration were also detected in HBe-positive and HBe-negative newly diagnosed patients, and although these cytokines increased in HBe-transfected U937 cells, serum IL-6 and IL-10 between HBe-positive and HBe-negative patients had no significant difference (IL-6: HBe positive vs. HBe negative: 2.69 ± 3.21 pg/mL vs. 0.78 ± 0.81 pg/mL, p = 0.1438; IL-10: HBe positive vs. HBe negative: 187.60 ± 91.12 vs. 254.59 ± 142.18 pg/mL, p = 0.1773; Fig. 2F).
HBe promoted B lymphocyte activation marker expression
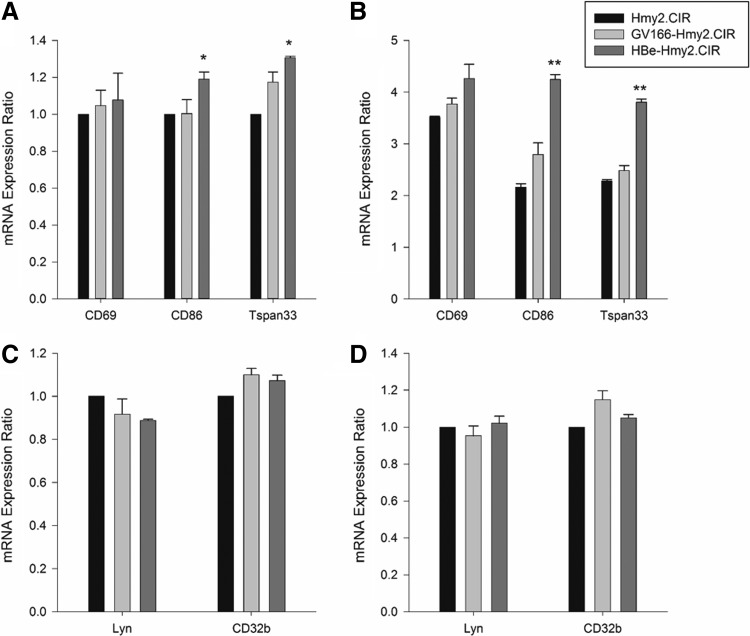
To further evaluate the activation status and the response to stimulus of HBe-infected B cells, semiquantitative RT-PCR was used to detect the expression of B cell activation markers CD69, CD86, and Tspan33, as well as the inhibiting regulation molecules Lyn and CD32b, in Hmy2.CIR and transfected Hmy2.CIR cells. We found that in the resting state, the expression of CD86 and Tspan33 was slightly increased in HBe-Hmy2.CIR (CD86: GV166-Hmy2.CIR vs. HBe-Hmy2.CIR: 1.00 ± 0.08 vs. 1.19 ± 0.04, p = 0.0324; Tspan33: GV166-Hmy2.CIR vs. HBe-Hmy2.CIR: 1.17 ± 0.06 vs. 1.31 ± 0.01, p = 0.0500; Fig. 4A). After being stimulated with CD40L and IL-4, all the expression of CD86 and Tspan33 increased by 2–3 times and maintained the similar difference trend with prior stimulation (CD86: GV166-Hmy2.CIR vs. HBe-Hmy2.CIR: 2.79 ± 0.22 vs. 4.25 ± 0.09, p = 0.0031; Tspan33: GV166-Hmy2.CIR vs. HBe-Hmy2.CIR: 2.48 ± 0.10 vs. 3.81 ± 0.06, p = 0.0001; Fig. 4B). These suggested that HBe infection may lead to B cell activation and increase B cell response to antigens. However, with or without stimulus, the expression of CD69 in HBe-Hmy2.CIR had no significant difference compared with control. As for Lyn and CD32b, the mRNA expression in each group was basically at the same level, regardless of whether IgM μ chain was added as stimulus (Fig. 4C, D). All of the above further prove that B cells were in a hyperactivated status in HBV infections.
FIG. 4.
B lymphocyte activation markers and inhibitory regulation molecules determined by semiquantitative RT-PCR. (A, B) mRNA expression of B cell activation markers CD69, CD86, and TSPAN33 in Hmy2.CIR and transfected Hmy2.CIR cells. (A) Without stimulation. This result showed that in the resting state, the expression of two B lymphocyte activation markers CD86 and TSPAN33 was slightly increased in HBe-Hmy2.CIR cells. (B) After stimulation with CD40L (1.5 μg/mL) and IL-4 (3 ng/mL) for 12 h. This result presented a significant increase of CD86 and TSPAN33 expression in HBe-Hmy2.CIR cells after these cells were activated. (C, D) mRNA expression of B cell inhibitory regulation molecules CD32b and Lyn in Hmy2.CIR and transfected Hmy2.CIR cells. (C) Without stimulation, (D) stimulated with IgM μ chain (2 μg/mL) for 8 h. No significant differences of these two inhibition markers of B lymphocytes were detected between HBe-Hmy2.CIR and GV166-Hmy2.CIR cells. All experiments were repeated thrice independently. Differences between groups were analyzed using the independent samples Student's t-test and considered to be significant if p < 0.05. *p < 0.05, **p < 0.01.
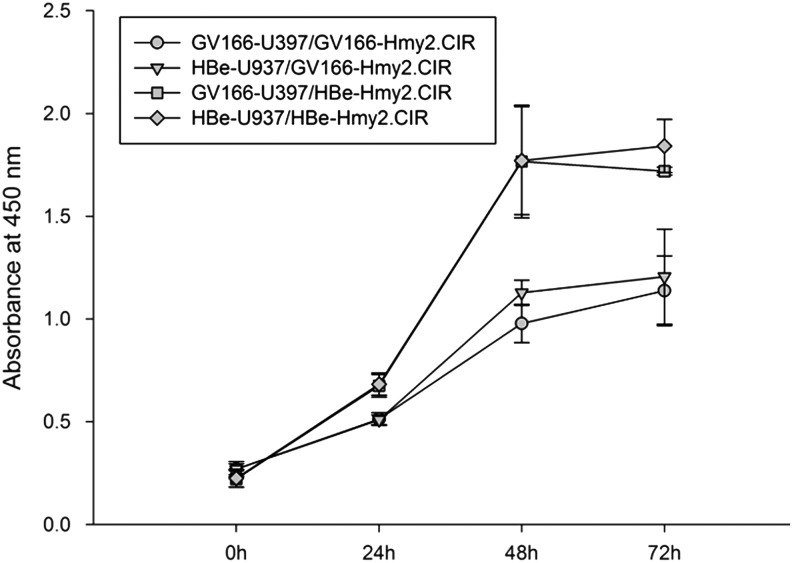
HBe transfection itself enhanced Hmy2.CIR proliferation
After coculture for indicated times, proliferation of GV166-Hmy2.CIR and HBe-Hmy2.CIR was detected with the Cell Counting Kit-8. Absorbance at 450 nm of each group was measured to reflect degrees of cell proliferation. Results showed that regardless of GV166-Hmy2.CIR and HBe-Hmy2.CIR, coculture with GV166-U937 or HBe-U937 had no significant influence on proliferation of these cells (Fig. 5). However, HBe-Hmy2.CIR proliferation was significantly increased compared with GV166-Hmy2.CIR whether cocultured with GV166-U937 or HBe-U937 (GV166-U937/GV166-Hmy2.CIR vs. GV166-U937/HBe-Hmy2.CIR: 24-h p-value: 0.0112, 48-h p-value: 0.0091, 72-h p-value: 0.0041; HBe-U937/GV166-Hmy2.CIR vs. HBe-U937/HBe-Hmy2.CIR: 24-h p-value: 0.0066, 48-h p-value: 0.0145, 72-h p-value: 0.0143; Fig. 5). These results emphasized that coculture with HBe-transfected U937 cells did not enhance Hmy2.CIR proliferation as expected, however, HBe transfection itself significantly promoted Hmy2.CIR proliferation.
FIG. 5.
HBe-positive and HBe-negative Hmy2.CIR proliferation in coculture systems with HBe-positive or HBe-negative U937. GV166-Hmy2.CIR and HBe-Hmy2.CIR were cocultured with GV166-U937 or HBe-U937 for indicated times. After that, proliferation of GV166-Hmy2.CIR and HBe-Hmy2.CIR was detected using Cell Counting Kit-8. Absorbance at 450 nm of each group was used to reflect cell proliferation. It showed that coculture with GV166-U937 or HBe-U937 did not influence GV166-Hmy2.CIR and HBe-Hmy2.CIR proliferation; however, HBe-Hmy2.CIR proliferation was significantly increased compared with GV166-Hmy2.CIR, whether cocultured with GV166-U937 or HBe-U937.
Discussion
HBe antigen is a nonstructural viral protein, which is a serum marker for HBV replication. It had been widely reported that, it has great immune regulation roles and associates with chronic hepatitis development (6,7,23–25). The presence of extrahepatic HBV DNA had been widely studied, and it had been proved that it existed in PBMCs from both acute and chronic hepatitis B (CHB) patients (18,31). However, their role in HBV infection and viral persistence remains unclear. In this study, we transfected U937 and Hmy2.CIR cell lines with HBe-contained lentivirus, and functions of these cells were determined. HBe DNA, mRNA, and HBe antigen were successfully detected in these transfected cells, which partly mimicked the process of HBe in HBV-infected monocytes and B lymphocytes. These models can reflect HBe infection functions on monocytes and B lymphocytes to some extent. However, U937 (histiocytic lymphoma cells) and Hmy2.CIR (B lymphoblast lymphomas cells) are different from primary monocytes and B lymphocytes in cell traits. In addition, lentivirus transfection itself will partly affect cell characters. Therefore, our research only aimed to reveal how HBe infection influences U937 and Hmy2.CIR functions, which may have some limitations for studying HBe infection roles in primary cells in HBV-infected patients.
Monocyte migration into infection cites plays critical roles in the clearance of pathogenic substances and inflammatory injury to the organism. Recently, it has been reported that HBe binds to monocytes, neutrophils, and B lymphocytes, inhibits monocyte motility, and suppresses the respiratory burst of monocytes and neutrophils (20). This result represents a negative regulatory role for HBe in monocytes, triggering innate immune response against HBV infection. In our study, cell migration of HBe-transfected U937 cells was sufficiently suppressed, which proved that HBe infection can inhibit U937 motility. Cytokine production in infection cites by monocytes is another important innate immunity response during infection. BAFF is an important B lymphocyte stimulation factor, which plays unique roles in B cell proliferation, differentiation, survival, selection, activation, and antibody production (11). There are three kinds of BAFF receptors in B lymphocyte. BAFF-R, also known as BlyS Receptor 3 (BR3), is the most affinity BAFF receptor (11). BAFF is produced primarily by cells of myeloid lineage, including monocytes, and it is initially synthesized in a membrane-anchored form and finally released as soluble BAFF after enzymatic cleavage (28). Our results showed that HBe-enhanced BAFF expression in U937, for the total BAFF mRNA level and soluble BAFF in culture supernatant, was all increased in HBe-U937 cells. Although BAFF production in HBe-transfected U937 was increased, we found that membrane-bound BAFF on HBe-U937 cells was decreased significantly compared with control, which indicated that BAFF release is promoted by HBe infection. This is, to the best of our knowledge, the first report addressing about HBe promoting BAFF release from membrane of monocytes. By further measurement of serum BAFF in HBe antigen-positive and HBe antigen-negative patients with HBV infection, we demonstrated that serum BAFF in HBe antigen-positive patients was higher than in HBe antigen-negative patients. This result confirms that HBe antigen promotes BAFF synthesis and release in vivo. In fact, BAFF levels in CHB patients have been determined previously and revealed that serum BAFF in CHB patients was higher than in healthy controls and is associated with clinical disease (35). BAFF-R expressed on HBe-Hmy2.CIR has no significant difference compared with GV166-Hmy2.CIR. This indicates that HBe does not influence B lymphocyte activation by regulating BAFF-R expression, but it may accelerate B cell activation by enhancing BAFF production and release by monocytes. Expression of another B lymphocyte activation factor APRIL in U937 and transfected U937 cells was basically the same.
IL-6 is a multifunctional proinflammatory cytokine that plays a great role in the response of hepatic epithelia to inflammation, and it has been proved to be involved in cancer progress, including HCC and cholangiocarcinoma (CCA) (13,14,17). In HBV-infected patients, IL-6 was significantly increased and the expression of IL-6 showed correlation with impaired liver function in naive hepatitis B infection (CHB) patients (15). IL-6 is primarily produced by monocytes and Kupffer cells in liver. It had been reported that expression of TLR2 on peripheral monocytes and Kupffer cells were reduced in HBe-positive CHB patients, and production of TNF-α and IL-6 in peripheral monocytes when stimulated with TLR2 ligand was reduced in vitro (33). However, in CHB patients, IL-6 expression had been proved to not be associated with HBe antigen (5,15). In our study, a remarkable decrease in IL-6 production in GV166-U937 and HBe-U937 compared with U937 was noticed, which indicated that lentivirus transfection itself sufficiently influenced U937 character in IL-6 production. This result reminds us of a limitation in using transfected U937 to study HBe function in monocytes in this research. However, when compared with the GV166-U937 group, IL-6 significantly increased in the HBe-U937 group after being stimulated with LPS, which reflected that HBe promoted IL-6 production stimulated by the LPS/TRL4 pathway in monocytes. Besides, our study confirmed that IL-6 expression in CHB patients was not associated with the HBe antigen status. Except for IL-6, another proinflammatory cytokine, TNF-α, detected in this article had no significant difference between groups. IL-10 is a kind of anti-inflammation cytokine, which is mainly produced by monocytes, and plays a role in triggering earlier adaptive cellular responses. Herein, like IL-6, IL-10 production in HBe-U937 cells was significantly increased compared with GV166-HBe. However, serum IL-10 between HBe-positive and HBe-negative CHB patients had no significant difference in our study, which is consistent with a previous report (29).
Neutralizing antibodies play important roles in the control of viremia, and the ability of human body to produce specific antibody-secreting B cells directly influences the outcome of virus infection (9,10). In addition, B cells can act as antigen-presenting cells (APCs) to present antigen through MHC class I and II pathways and stimulate HBV-specific CD4 and CD8 T cell responses (16,19,22). Therefore, B lymphocyte activation status may involve in immune response to HBV infection. CD69 is an early activation molecule on B lymphocytes and CD86 is a costimulatory molecule for T cell receptor. Some researchers had used these two activation markers to detect the activation status of peripheral blood B lymphocytes before and they confirmed that B cell activation was stronger in patients with HBV infection than in controls and related to the severity of disease (34). In this study, we failed to detect excessive expression of CD69 in HBe-transfected Hmy2.CIR cells, but CD86 expression was elevated in HBe-transfected Hmy2.CIR cells. CD86 is a costimulatory molecule expressed on APCs, providing important costimulatory signal for T cell activation (32). Exorbitant expression of CD86 on HBe-transfected B lymphocytes presumed that these cells were more sensitive for stimulus or antigens, and had a higher capability to activate T cells as APCs. Tspan33 is expressed only in malignant B cells and the activated B cell, and it may be involved in B cell activation, maturation, survival, or antigen presentation (21). Tspan33 expression was increased in HBe-transfected Hmy2.CIR cells, further confirmed a higher activation state of B lymphocytes when influenced by HBe. A variety of studies have highlighted the role of inhibitory receptors in setting a threshold for B cell activation. To comprehensively analyze B cell physiological and antibody production in HBV infection, we detected the expression of inhibitory receptors in Hym2.CIR cells. Two inhibitory markers Lyn and CD32b were measured. Lyn is a crucial mediator of inhibitory signaling pathways in B cells and normally counterbalances B cell antigen receptor (BCR) signaling pathway by phosphorylating several inhibitory immunoglobulin receptors in B cells such that B cell differentiation is limited. CD32b, known as FcγRIIB, is one of the exact targets of Lyn and downregulates the BCR signaling complex. CD32b expresses at all development stages of B cells from pre-B cell to memory B cells, and plasma cells (4). However, in this study, no evidence for defective inhibitory signaling was found in HBe-transfected B cells before and after stimulation. This result indicates that B lymphocyte activation threshold was not influenced by HBe.
HBe-Hmy2.CIR cells showed activated phenotypes in CD86 and Tspan33 as mentioned above, which indicated an intensive activation status of these cells. Hence, we further determined proliferations of GV166-Hmy2.CIR and HBe-Hmy2.CIR cells after coculture with GV166-U937 or HBe-U937. It showed that GV166-Hmy2.CIR or HBe-Hmy2.CIR cocultured with HBe-positive U937 (HBe-U937) would not influence their proliferations. These results may be due to the limitation of cell numbers loaded in the upper inserts in the coculture systems. Although expression of BAFF, IL-6, and IL-10 was proven higher in HBe-positive U937 cells, the tiny difference caused by insufficient GV166-U937 or HBe-U937 loaded in the coculture systems could not influence proliferation of transfected Hmy2.CIR. However, HBe-Hmy2.CIR proliferation was significantly increased compared with GV166-Hmy2.CIR whether cocultured with GV166-U937 or HBe-U937. These demonstrated that HBe infection itself sufficiently increased Hmy2.CIR proliferation.
In conclusion, our results demonstrate that HBe inhibits U937 migration. HBe enhances BAFF production and leads to BAFF release markedly increase in U937. Meanwhile, HBe promotes IL-6 and IL-10 expression in U937 with LPS stimulation. Hmy2.CIR activation status is dramatically elevated with the regulation of HBe, and HBe infection promotes Hmy2.CIR proliferation. These indicate that HBe can enhance the immune activeness of B cells, both through promoting BAFF production and direct induction.
Acknowledgments
This study was supported by Shandong Provincial Natural Science Foundation (Grant No. ZR2012HM054) and the National Natural Science Funds of China (Grant No. 31370897). We thank all participants in the study for making this work possible.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Ait-Goughoulte M, Lucifora J, Zoulim F, et al. Innate antiviral immune responses to hepatitis B virus. Viruses 2010;2:1394–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberti A, Diana S, Sculard GH, et al. Detection of a new antibody system reacting with Dane particles in hepatitis B virus infection. Br Med J 1978;2:1056–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachmann MF, and Kopf M. The role of B cells in acute and chronic infections. Curr Opin Immunol 1999;11:332–339 [DOI] [PubMed] [Google Scholar]
- 4.Baerenwaldt A, Lux A, Danzer H, et al. Fcgamma receptor IIB (FcgammaRIIB) maintains humoral tolerance in the human immune system in vivo. Proc Natl Acad Sci U S A 2011;108:18772–18777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boltjes A, Groothuismink ZM, van Oord GW, et al. Monocytes from chronic HBV patients react in vitro to HBsAg and TLR by producing cytokines irrespective of stage of disease. PLoS One 2014;9:e97006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunetto MR, Giarin MM, Oliveri F, et al. Wild-type and e antigen-minus hepatitis B viruses and course of chronic hepatitis. Proc Natl Acad Sci U S A 1991;88:4186–4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen MT, Billaud JN, Sallberg M, et al. A function of the hepatitis B virus precore protein is to regulate the immune response to the core antigen. Proc Natl Acad Sci U S A 2004;101:14913–14918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chisari FV, and Ferrari C. Hepatitis B virus immunopathogenesis. Annu Rev Immunol 1995;13:29–60 [DOI] [PubMed] [Google Scholar]
- 9.Ciurea A, Hunziker L, Klenerman P, et al. Impairment of CD4(+) T cell responses during chronic virus infection prevents neutralizing antibody responses against virus escape mutants. J Exp Med 2001;193:297–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciurea A, Klenerman P, Hunziker L, et al. Viral persistence in vivo through selection of neutralizing antibody-escape variants. Proc Natl Acad Sci U S A 2000;97:2749–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goenka R, Scholz JL, Sindhava VJ, et al. New roles for the BLyS/BAFF family in antigen-experienced B cell niches. Cytokine Growth Factor Rev 2014;25:107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grady GF, Lee VA, Prince AM, et al. Hepatitis B immune globulin for accidental exposures among medical personnel: final report of a multicenter controlled trial. J Infect Dis 1978;138:625–638 [DOI] [PubMed] [Google Scholar]
- 13.He G, and Karin M. NF-kappaB and STAT3-key players in liver inflammation and cancer. Cell Res 2011;21:159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson C, Han Y, Hughart N, et al. Interleukin-6 and its receptor, key players in hepatobiliary inflammation and cancer. Transl Gastrointest Cancer 2012;1:58–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kao JT, Lai HC, Tsai SM, et al. Rather than interleukin-27, interleukin-6 expresses positive correlation with liver severity in naive hepatitis B infection patients. Liver Int 2012;32:928–936 [DOI] [PubMed] [Google Scholar]
- 16.Ke Y, and Kapp JA. Exogenous antigens gain access to the major histocompatibility complex class I processing pathway in B cells by receptor-mediated uptake. J Exp Med 1996;184:1179–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong L, Zhou Y, Bu H, et al. Deletion of interleukin-6 in monocytes/macrophages suppresses the initiation of hepatocellular carcinoma in mice. J Exp Clin Cancer Res 2016;35:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laskus T, Radkowski M, Wang LF, et al. Detection and sequence analysis of hepatitis B virus integration in peripheral blood mononuclear cells. J Virol 1999;73:1235–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazdina U, Alheim M, Nystrom J, et al. Priming of cytotoxic T cell responses to exogenous hepatitis B virus core antigen is B cell dependent. J Gen Virol 2003;84:139–146 [DOI] [PubMed] [Google Scholar]
- 20.Leu CM, Lu YC, Peng WL, et al. The hepatitis B virus e antigen suppresses the respiratory burst and mobility of human monocytes and neutrophils. Immunobiology 2014;219:880–887 [DOI] [PubMed] [Google Scholar]
- 21.Luu VP, Hevezi P, Vences-Catalan F, et al. TSPAN33 is a novel marker of activated and malignant B cells. Clin Immunol 2013;149:388–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milich DR, Chen M, Schodel F, et al. Role of B cells in antigen presentation of the hepatitis B core. Proc Natl Acad Sci U S A 1997;94:14648–14653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milich DR, Chen MK, Hughes JL, et al. The secreted hepatitis B precore antigen can modulate the immune response to the nucleocapsid: a mechanism for persistence. J Immunol 1998;160:2013–2021 [PubMed] [Google Scholar]
- 24.Milich DR, Jones JE, Hughes JL, et al. Is a function of the secreted hepatitis B e antigen to induce immunologic tolerance in utero? Proc Natl Acad Sci U S A 1990;87:6599–6603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milich DR, Schodel F, Hughes JL, et al. The hepatitis B virus core and e antigens elicit different Th cell subsets: antigen structure can affect Th cell phenotype. J Virol 1997;71:2192–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore PA, Belvedere O, Orr A, et al. BLyS: member of the tumor necrosis factor family and B lymphocyte stimulator. Science 1999;285:260–263 [DOI] [PubMed] [Google Scholar]
- 27.Murakami Y, Minami M, Daimon Y, et al. Hepatitis B virus DNA in liver, serum, and peripheral blood mononuclear cells after the clearance of serum hepatitis B virus surface antigen. J Med Virol 2004;72:203–214 [DOI] [PubMed] [Google Scholar]
- 28.Nardelli B, Belvedere O, Roschke V, et al. Synthesis and release of B-lymphocyte stimulator from myeloid cells. Blood 2001;97:198–204 [DOI] [PubMed] [Google Scholar]
- 29.Ozguler M, Akbulut HH, and Akbulut A. Evaluation of interleukin-10 levels in patients diagnosed with chronic hepatitis. West Indian Med J 2015;64:71–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seeger C, and Mason WS. Molecular biology of hepatitis B virus infection. Virology 2015;479–480:672–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trippler M, Meyer zum Buschenfelde KH, and Gerken G. HBV viral load within subpopulations of peripheral blood mononuclear cells in HBV infection using limiting dilution PCR. J Virol Methods 1999;78:129–147 [DOI] [PubMed] [Google Scholar]
- 32.Vasilevko V, Ghochikyan A, Holterman MJ, et al. CD80 (B7-1) and CD86 (B7-2) are functionally equivalent in the initiation and maintenance of CD4+ T-cell proliferation after activation with suboptimal doses of PHA. DNA Cell Biol 2002;21:137–149 [DOI] [PubMed] [Google Scholar]
- 33.Visvanathan K, Skinner NA, Thompson AJ, et al. Regulation of Toll-like receptor-2 expression in chronic hepatitis B by the precore protein. Hepatology 2007;45:102–110 [DOI] [PubMed] [Google Scholar]
- 34.Wang K, Pei H, Huang B, et al. Overexpression of Fc receptor-like 1 associated with B-cell activation during hepatitis B virus infection. Braz J Med Biol Res 2012;45:1112–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang C, Li N, Wang Y, et al. Serum levels of B-cell activating factor in chronic hepatitis B virus infection: association with clinical diseases. J Interferon Cytokine Res 2014;34:787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]