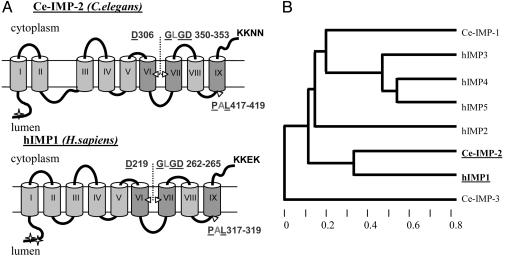
Fig. 1.
Similarity of Ce-IMP-2 to other members of IMPAS and presenilin families of proteins. (A) Predicted transmembrane structure of Ce-IMP-2, C. elegans, and hIMP1,Homo sapiens. The arrowheads denote the positions of conserved aspartate and PAL-motif residues. The letters show invariant amino acid residues in eukaryotic PSs and IMPASes; the underlined letters are identical also in related proteins from Archaea and bacterial polytopic type 4 prepilin peptidases (TFPP). Most conserved domains in PSs and IMPAS families are shown in intense gray. The predicted sites of N-glycosylation (marked by asterisks) and endoplasmic reticulum-membrane retention signals (KKXX motif) are shown (netnglyc 1.0 and psort ii predictions). (B) Phylogenic tree (Cluster algorithm, genebee) for conserved domains (amino acid 269-444) of C. elegans Ce-IMP-2 and corresponding domains of C. elegans Ce-IMP-1, Ce-IMP-3, and human IMP (hIMP1-5) proteins.