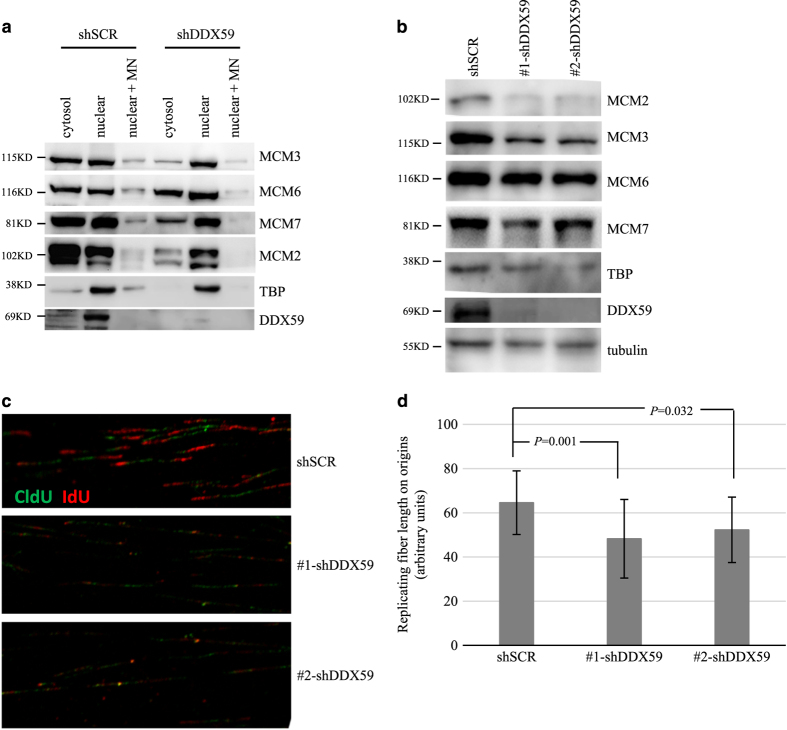
Figure 6.
DDX59 knockdown reduces chromatin bound and total levels of MCM proteins. (a) H1299 cells were transduced with either shSCR or shRNA-DDX59 lentivirus. Four days post infection, cells were collected for cytosolic, nuclear soluble and chromatin-bound (by micrococcal nuclease treatment) protein fractionations. Equal proportion of each fractionation was then analyzed by western blot with the indicated antibodies. (b) H1299 cells were transduced with either shSCR or shRNA-DDX59 lentivirus. Four days post infection, cells were collected for total protein analysis with the indicated antibodies. (c) Above-mentioned cells were pulse-labeled with 50 μM CldU (15 min) followed by 250 μM IdU (15 min). DNA fibers were spreaded from labeled cells. Each labeled fork in the replication origins was shown as –red–green–red– segment. Typical images were shown for each sample. (d) The length for each replication fork was calculated from 100 tracks per condition. Data shown were the averages with S.d. bars for up to 100 tracks. P was calculated and shown, statistically significant difference was observed after DDX59 knockdown as compared with the shSCR control.