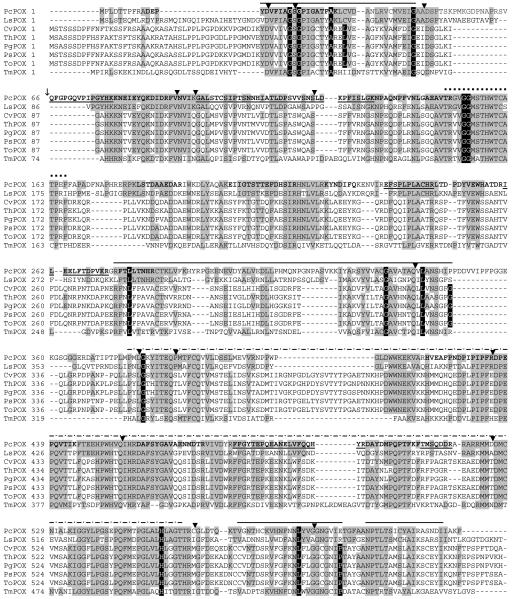
FIG. 2.
Alignment of P. chrysosporium BKM-F-1767 POX (PcPOX; accession no. AY522922) translated from cDNA sequence, with the POXs from Lyophyllum shimeji (LsPOX; accession no. gi|44886072|), Coriolus versicolor (CvPOX; accession no. gi|25091018| [33]), T. hirsuta (ThPOX; accession no. gi|25091016| [7]), Peniophora gigantea (PgPOX; accession no. gi|34452037|), Peniophora sp. (PsPOX; accession no. gi|27436422|), Trametes ochracea (ToPOX; accession no. gi|31044224|) and Tricholoma matsutake (TmPOX; accession no. gi|25553433| [44]). Grey shaded amino acids indicate homology with PcPOX; black shaded amino acids with white typeface indicate CvPOX-GMC consensus sequences (see the text). Solid lines above sequences indicate FAD-binding domains; dotted lines above sequences indicate flavin attachment loops; dashed lines above sequences indicate substrate binding domains. Boldface indicates peptides identified by LC-MS/MS; underlined and boldface type distinguishes separate peptides within a continuous sequence. ▾, intron positions; ↓, the start of the N terminus reported by Artolozaga et al. (2) for the POX of P. chrysosporium K3.