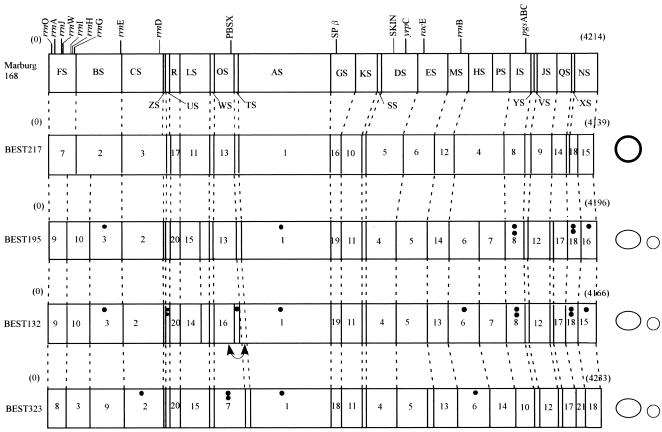
FIG. 5.
Physical maps of B. subtilis (natto) strains. The map of Marburg 168, linearized at the SfiI site between FS and NS closest to the origin of replication (oriC), is shown at the top. Similar linearized forms of SfiI maps for the four natto strains are shown below the Marburg 168 map. Plasmids that have no SfiI site are shown on the right. Large plasmids are represented by large ovals, and small plasmids are represented by small circles. pL32 from BEST217 is represented by a boldface circle to distinguish it from the pLS20 family plasmid represented by an oval. Plasmid sizes are not to scale. Locations of the 10 rrn operons containing the sites for I-CeuI, prophages, PBSX, SPβ, and skin are shown on the Marburg 168 map. Genes mapped in this study are also indicated on the Marburg 168 map. The number of dots indicates the number of IS determined from the data in Fig. 6. A small inversion was suggested in the BEST132 genome. This inversion, indicated by a double-headed curved arrow, is based on the Southern hybridization pattern for the two adjacent linking clones (pTA-168 and pOT-168), which was different from that of other strains. The order of hybridized bands with these two linking clones for BEST132 was reversed.