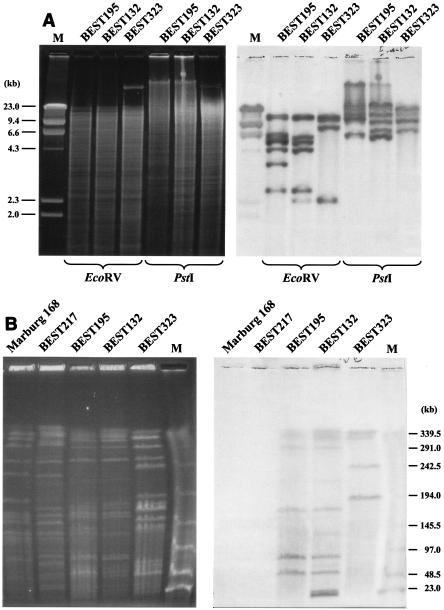
FIG. 6.
Analysis of IS elements in the B. subtilis (natto) genome. (A) Estimation of the copy number of IS4Bsu1. EcoRV and PstI digests of genomic DNA of three strains were subjected to agarose gel electrophoresis (left gel) and hybridized with the IS4Bsu1 probe (right gel). The number of IS copies per genome is equivalent to the number of hybridized bands because there are no EcoRV and PstI sites in the IS4Bsu1sequence. Lane M contained lambda phage DNA as a size marker. (B) Mapping on SfiI fragments with multiple copies of the IS element. The SfiI digests of genomic DNA of strains were resolved by using a contour-clamped homogeneous electric field with the following running conditions: 3 V/cm, a pulse time of 24 s, and a running time of 56 h (left gel). The blot was hybridized (right gel) with IS4Bsu1 as a probe (25). The intensities of the Southern signal were digitally treated, and number of a particular SfiI fragment was calculated based on the total number per genome derived from the data in panel A. The results are shown in Fig. 5. Lane M contained lambda phage DNA as a size marker.