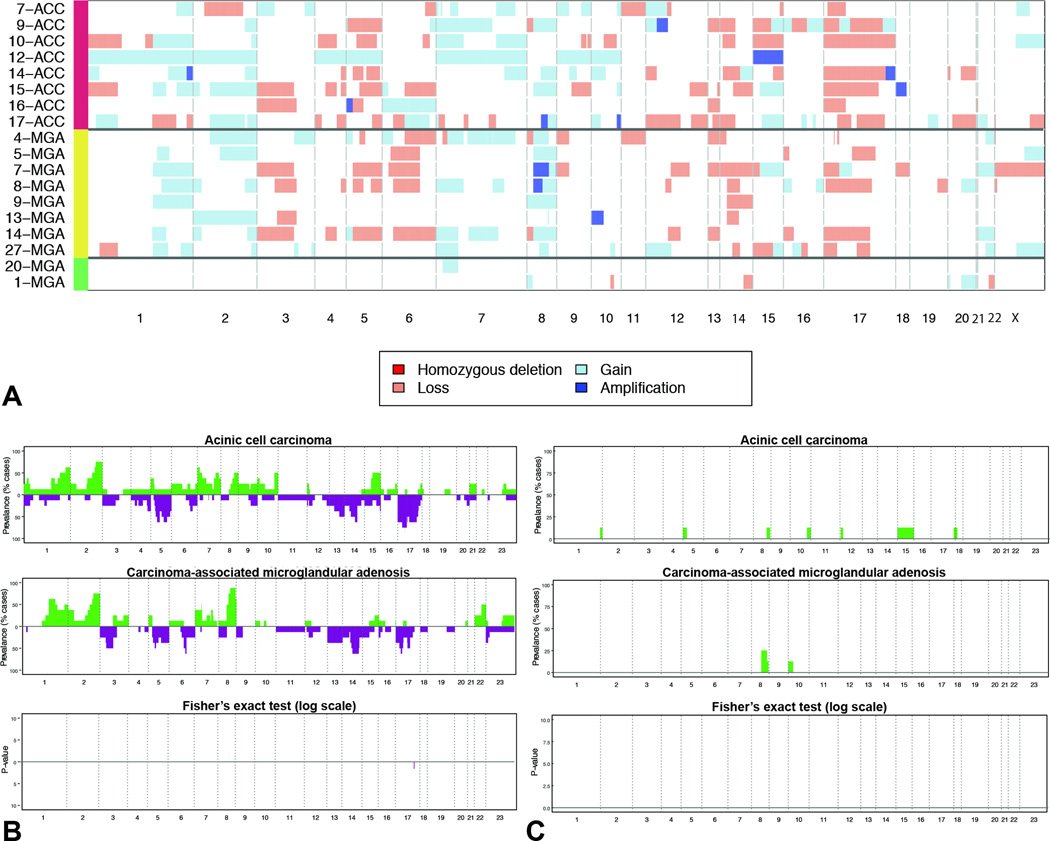
Figure 3. Repertoire of copy number alterations identified in microglandular adenoses and acinic cell carcinomas.
A) Heatmap depicting the copy number alterations identified in the pure microglandular adenoses (n=2), carcinoma-associated microglandular adenoses/atypical microglandular adenoses (n=8) and acinic cell carcinomas (n=8) analyzed. Samples are represented on the y-axis, copy number alterations are represented along the x-axis according to their respective genomic location. Light red: copy number loss; white: neutral; light blue: copy number gain; dark blue: amplification. B) Frequency plots of recurrent gains and losses in acinic cell carcinomas (top) and carcinoma-associated microglandular adenoses/atypical microglandular adenoses (middle). Significant differences (Fishers’ exact test, p<0.05) are plotted in the bottom panel. On the y-axis the proportion of samples in which gains (green bars) or losses (purple bars) were identified is plotted according to genomic location (x-axis). C) Frequency plots of recurrent amplifications in acinic cell carcinomas (top) and carcinoma-associated microglandular adenoses/atypical microglandular adenoses (middle). Significant differences (Fishers’ exact test, p<0.05) are plotted in the bottom panel. On the y-axis the proportion of samples in which amplifications (green bars) were identified is plotted according to genomic location (x-axis).