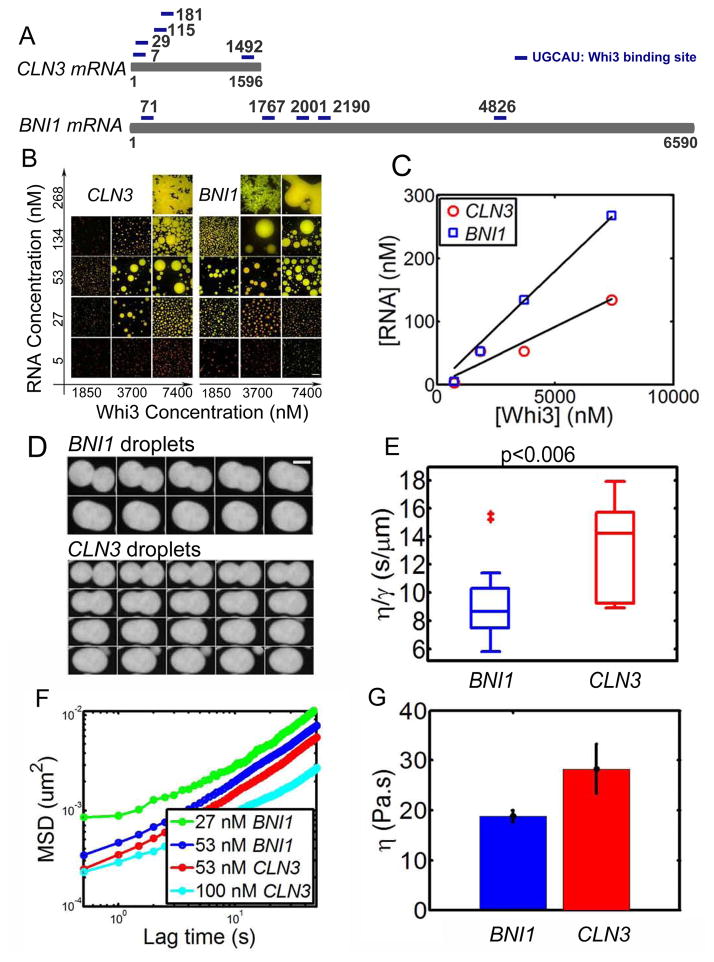
Figure 5. BNI1 mRNA drives Whi3 phase separate into droplets with different properties.
(A) Schematics show CLN3 mRNA and BNI1 mRNA, each with five Whi3 binding sites. (B) Whi3 (green) phase diagram with BNI1 mRNA (red) and CLN3 RNA (red), images were taken after 4 hours of mixing Whi3 with RNA at 150 mM salt. Scale bar is 20 μm. (C) RNA concentration at which the largest apparent droplet volume is observed in B for each Whi3 concentration. The optimal RNA/Whi3 molar ratio estimated from linear fit (black line) for CLN3 is ~0.02 (similar to that obtained from overnight droplets for CLN3 in Fig. S3) and BNI1 is ~0.04. (D) Example of fusion images for 50 nM CLN3 and BNI1 RNA with 9 μM Whi3 at150 mM salt shows a faster fusion rate for BNI1. Time interval between images is 10 second. Scale bar is 5 μm. (E) Box plot of viscosity to surface tension ratio (η/γ) for CLN3 and BNI1 droplets obtained from fusion events. (F) Mean MSD from microrheology for CLN3 and BNI1 with 8 μM Whi3 at 150 mM salt, showing faster movement of tracer beads for less RNA both for BNI1 and CLN3 droplets. (G) Viscosity of 53 nM BNI1 and CLN3 droplets obtained from MSD data, showing BNI1 droplets are less viscous. Mean±SEM.