FIGURE 4:
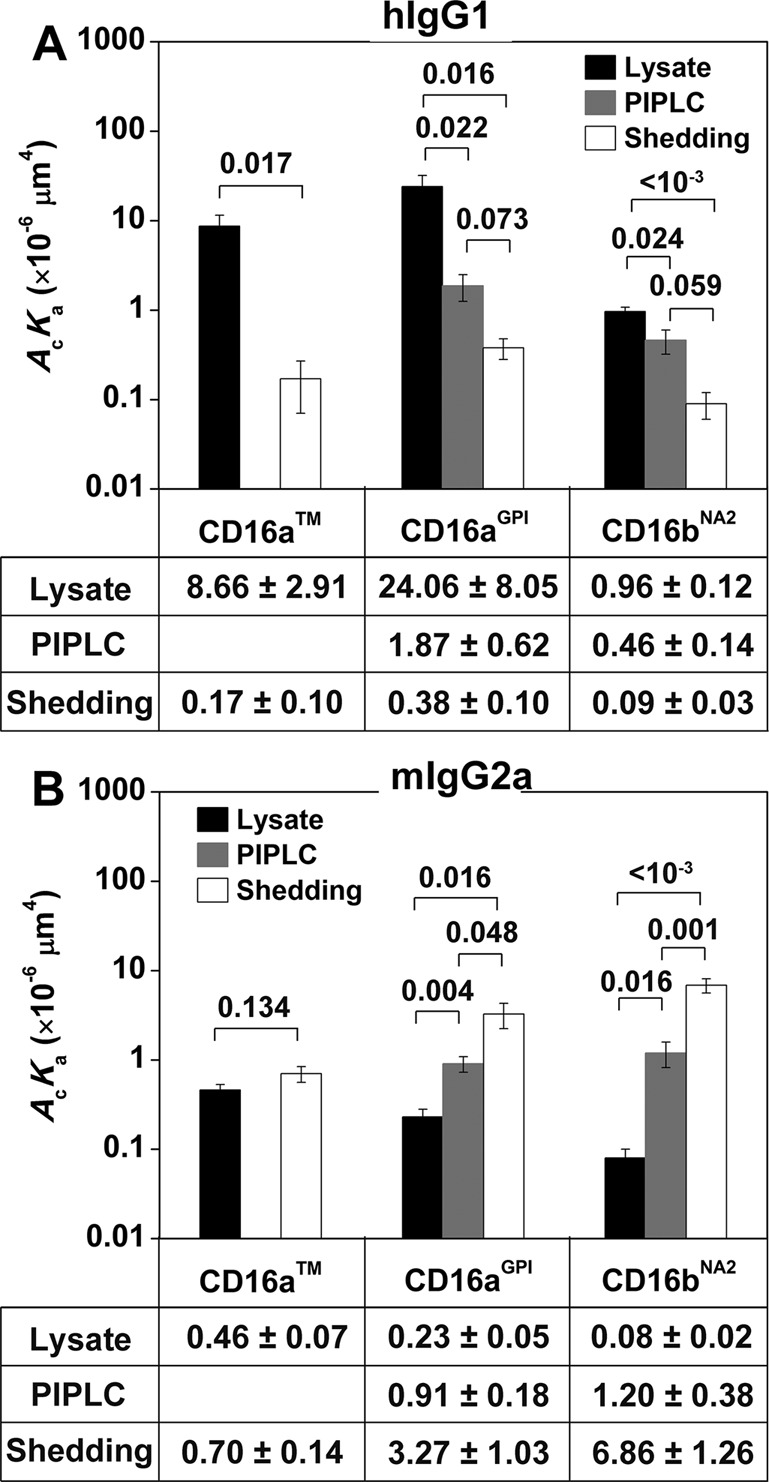
Comparisons of 2D affinities of hIgG1 (A) and mIgG2a (B) for CD16. CD16 molecules are captured on 214.1-precoated microspheres from CHO cell lysates, supernatants of CHO cells treated with PIPLC, or supernatants of CHO cells subjected to shedding treatment. Adhesion frequencies were measured with an 8-s contact duration and converted to 2D effective affinities using Eq. 3. Data are mean ± SEM of 5–15 RBC–microsphere pairs with 100 contacts each per bar. Data values are below the plots; p values from Student’s t test are above data bars.
