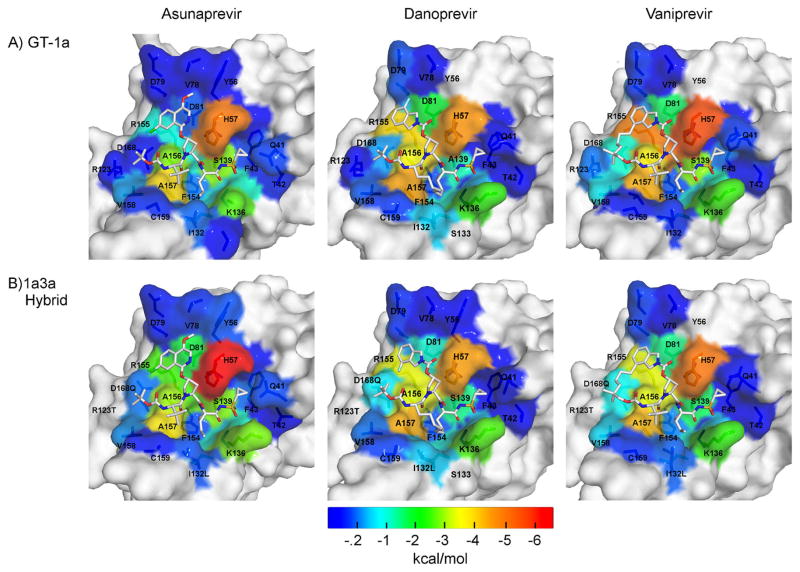
Figure 3.
Packing of inhibitors at the NS3/4A protease active site. The van der Waals (vdW) contact potentials, averaged from the MD simulations, of protease active site residues with the inhibitor: (A) GT-1 and (B) chimeric 1a3a construct complexes. The protease residues are colored blue to red for increasing contacts with the inhibitor mapped onto the protease surface of the cocrystal structures (1a/1a3a): ASV (4WF8/5EQS), DAN (3M5L/5EGR) and VAN (3SU3/5ESB). DAN’s isoindoline group is flipped between the two cocrystal structures.