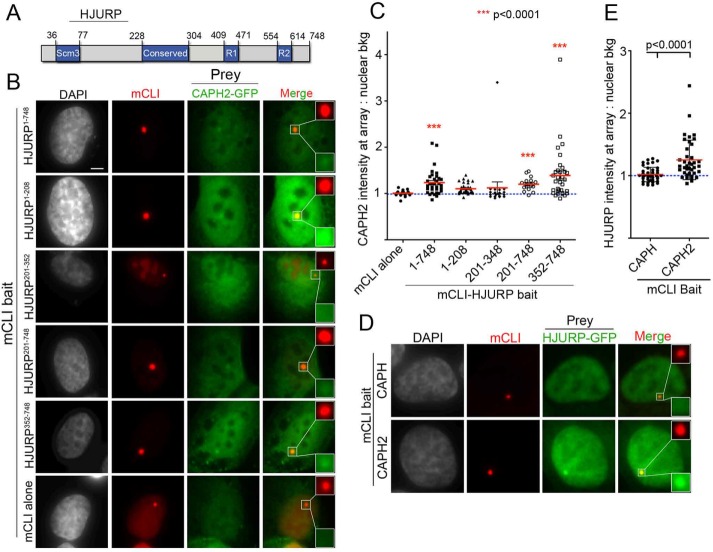
FIGURE 4:
HJURP interacts with condensin II via its C-terminal amino acids 352–748. (A) Schematic of HJURP and its known domains. Numbers are amino acids. (B) Representative images of U2OS-LacO cells cotransfected for 48 h with indicated mCLI-HJURP fragments and CAPH2-GFP. Cells were fixed without preextraction. Scale bar, 5 μm. (C) Quantification of experiment in B. Graph displays intensity of CAPH2-GFP at individual mCLI-HJURP arrays as a ratio over the average nuclear background. Red bar marks the mean, and whiskers mark the SD for each condition; ≥30 arrays/condition. ***p < 0.0001 when compared with mCLI by Kruskal–Wallis test followed by Dunn’s multiple comparison test. Blue dotted line marks an intensity ratio of 1, indicating no array enrichment. (D) Representative images of U2OS-LacO cells cotransfected for 48 h with mCLI-CAPH or CAPH2 as bait and HJURP-GFP as prey. Cells were fixed without preextraction. (E) Quantification of experiment in D. Graph displays intensity of HJURP-GFP at individual mCLI-CAPH or CAPH2 arrays as a ratio over the average nuclear background. Red bar marks the mean, and whiskers mark the SD for each condition; ≥20 arrays/condition, two biological replicates. p < 0.0001 by Mann–Whitney test. Blue dotted line marks an intensity ratio of 1, indicating no array enrichment.