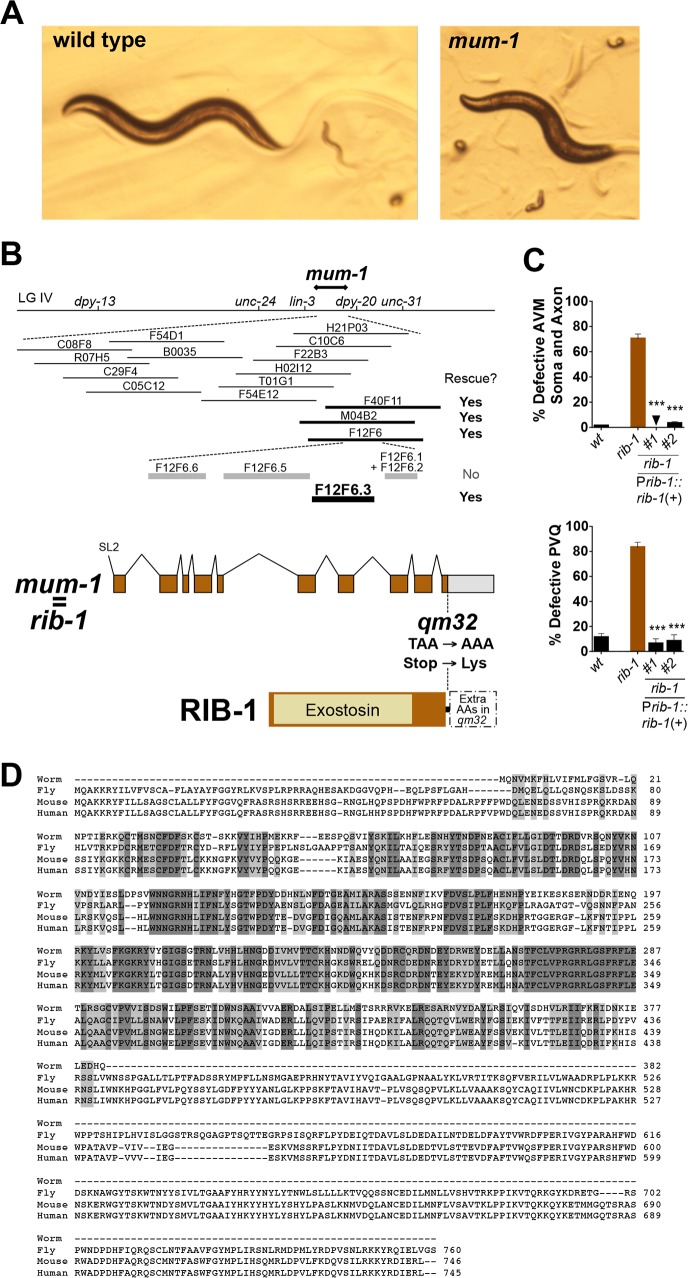
Fig 1. mum-1 is a viable hypomorphic mutation of the gene rib-1, which encodes one of the two HS copolymerase subunits.
A. Pictures of wild type and mum-1/rib-1 mutant animals. Some of the mum-1/rib-1 mutants die as embryos or misshapen larvae, and 14% of the animals reach adulthood and display defective locomotion and egg-laying, as described in [54]. B. Molecular identification of mum-1(qm32) as an allele of the gene rib-1. mum-1 was previously mapped between dpy-13 and unc-31 on linkage group IV [54]. We narrowed down its genetic position by a combination of three-point and two-point mapping (results were unc-24 70/79 mum-1 9/79 dpy-20; 10 Dpy-20 non Mum-1/4320 F2s; and lin-3 6/7 mum-1 1/7 dpy-20; which placed mum-1 between lin-3 and dpy-20, between 4.98 and 5.07 cM, according to the 95% confidence intervals). Cosmids (thin lines) and PCR products (thick lines) encompassing the gene rib-1(+) rescued the morphological and uncoordination defects of the mum-1 mutants. mum-1(qm32) is a missense mutation at base 39528 of cosmid F12F6 and changes the Stop into a Lys codon, which leads to a putative extension of 114 amino acids until the first in frame Stop codon. C. Rescue of the cell and axon guidance defects of the neuron AVM and axon guidance of the neurons PVQ of mum-1(qm32) with DNA corresponding to the genomic region of rib-1(+) (plasmid Prib-1::rib-1::Venus). See S2 and S3 Tables for sample sizes. Error bars are standard error of the proportion. Asterisks denote significant difference: *** P ≤ 0.001 (z-tests, P values were corrected by multiplying by the number of comparisons). D. An alignment of the predicted amino acid sequences of C. elegans RIB-1 (NP_502180.1) and its homologues from D. melanogaster (Ttv, NP_477231.1), M. musculus (EXT1, NP_034292.2), and H. sapiens (EXT1, NP_000118.2).