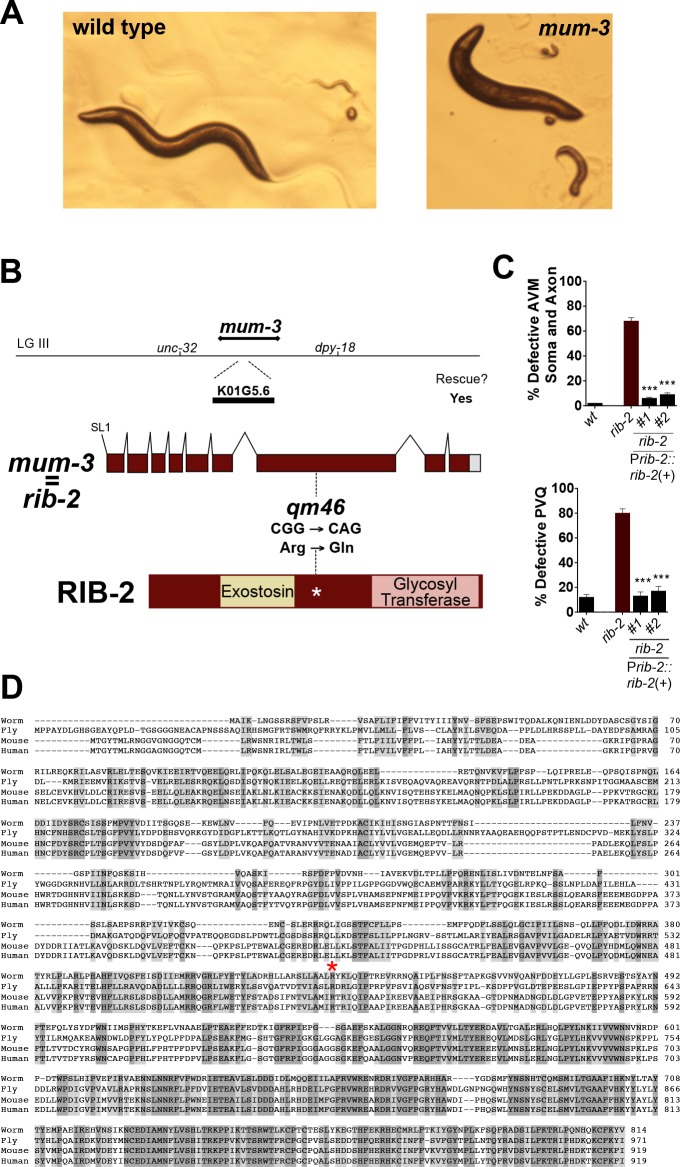
Fig 2. mum-3(qm46) is a viable hypomorphic mutation of the gene rib-2, which encodes the second HS copolymerase subunit.
A. Pictures of wild type and mum-3/rib-2 mutant worms. Some of the mum-3/rib-2 mutants die as embryos or deformed larvae, and 63% animals reach adulthood and display defective locomotion and egg-laying, as described in [54]. B. Molecular identification of mum-3(qm46) as an allele of the gene rib-2. mum-3 was previously mapped on linkage group III, between unc-32 and dpy-18 [54], which includes the second HSPG copolymerase gene rib-2. A PCR product containing the genomic locus of rib-2(+) rescued the morphological and uncoordination defects of the mum-3(qm46) mutants. mum-3(qm46) is a missense mutation at base 4366 on cosmid K01G5 that changes an Arg to a Gln at conserved residue 434, indicated by a red asterisk on the alignment (in D). C. Rescue of the cell and axon guidance defects of the neuron AVM and axon guidance of the neurons PVQ of mum-3(qm46) with DNA containing the genomic region of rib-2(+) (PCR product Prib-2::rib-2). See S2 and S3 Tables for sample sizes. Error bars are standard error of the proportion. Asterisks denote significant difference: *** P ≤ 0.001 (z-tests, P values were corrected by multiplying by the number of comparisons). D. An alignment of the predicted amino acid sequence of C. elegans RIB-2 (NP_499368.1) and its homologues from D. melanogaster (Botv, NP_523790.1), M. musculus (EXTL3, NP_061258.2), and H. sapiens (EXTL3, NP_001431.1).