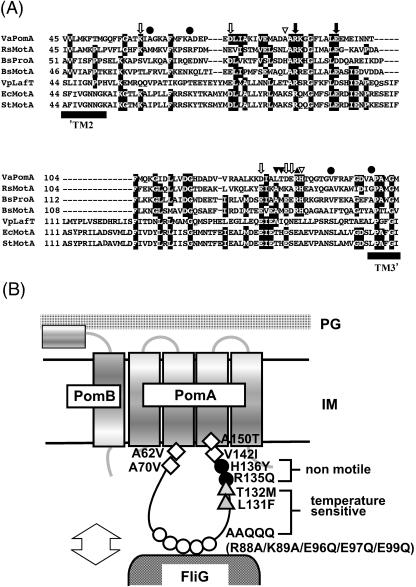
FIG. 1.
(A) Alignments of the PomA cytoplasmic loop with homologous proteins of various species. VaPomA, V. alginolyticus PomA; Rs MotA, R. sphaeroides MotA; BsProA, B. subtilis hypothetical protein A; BsMotA, B. subtilis MotA; VpLafT, V. parahaemolyticus LafT; EcMotA, E. coli MotA; StMotA, S. enterica serovar Typhimurium MotA. The residues are shown by white letters in black boxes when more than four identical residues are present at the aligned position. Black arrows indicate the two conserved charged residues. White arrows indicate the charged residues which were changed to neutral residues in this study. Black circles indicate the suppressor mutations of the TS PomA mutant. The paired black triangles and black or white inverted triangles show the sites of the previously isolated double mutations. (B) Mutations mapped in a schematic diagram of motor. ○, substitutions of AAQQQ (R88A, K89A, E96Q, E97Q, and E99Q); •, nonmotile mutations; ▵, substitutions required for the TS phenotype in addition to the AAQQQ substitutions; □, suppressor mutations of the TS mutant.