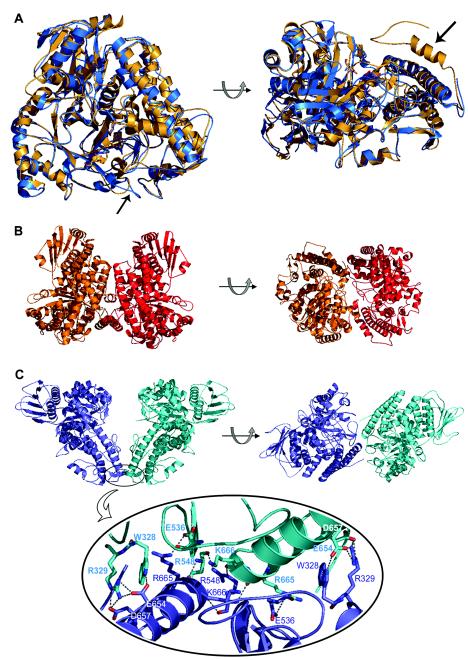
FIG.3.
(A) Superposition of the crystal structures of the α-glucuronidases AguA from G. stearothermophilus (blue) and GlcA67A from C. japonicus (gold), shown in two orientations. The two arrows point to the structural elements that are proposed to be responsible for the differences in the oligomerization of the two enzymes: the loop containing residues 323 to 329 in AguA (left) and the additional 29-residue polypeptide at the C terminus of GlcA67A (right). Superimposition was performed by the K2 server (29). (B) Two views of the GlcA67A dimer related by a 90° rotation (protein data bank code 1GQI). (C) Two views of the suggested AguA dimer related by a 90° rotation (protein data bank code 1L8N) (top). The lower part of the panel is an enlargement of the dimer interface region of AguA, showing the hydrogen bonds between the two subunits. The figures were generated by PyMOL (10).