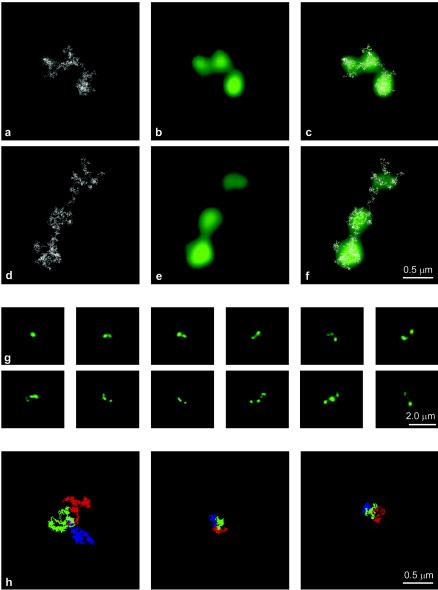
FIG. 5.
A NARC model can explain beaded images. (a and d) Examples of 400-kb simulated nucleosome chains. (b and e) High-magnification views of the simulated images reveal the presence of fluorescence puncta; most details of the nucleosome chain are lost. (c and f) Overlays show that less densely packed regions of the chain may extend beyond the puncta and may be invisible. (g) Sampling of simulated projected images following deconvolution for 400-kb chains with different degrees of decondensation. Note that these images exhibit beads resembling those in the actual images (compare to Fig. 3). (h) When the first third of the chain is colored blue, the middle third is colored green, and the last third is colored red, distinct largely nonoverlapping regions are obtained either for long chains of 256 kb (left panel) or for shorter chains of 64 kb (middle and right panels). These data indicate that, on a macroscopic scale, neighbor-neighbor relations are preserved, and so the chain could unravel without significant entanglement.