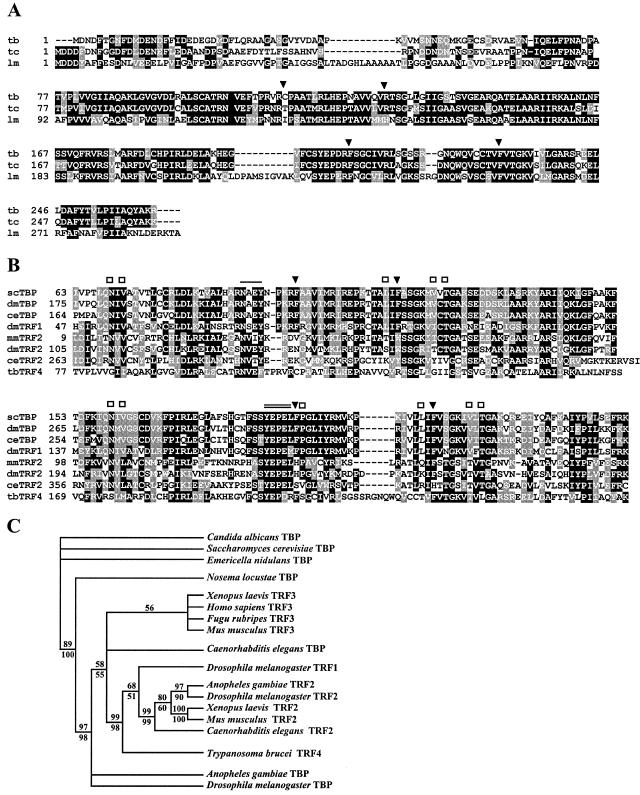
FIG. 1.
(A) ClustalW alignment of T. brucei TRF4 (tb; GeneDB accession no. Tb10.61.0330), T. cruzi TRF4 (tc; GeneDB accession no. Tc00.1047053503809.150) (note that the TRF4 ORF is encoded in the −1 reading frame), and L. major TRF4 (lm; GeneDB accession no. LmjF19.1390). Conserved amino acids (white letters on black background) and similar amino acids (white letters on shaded background) are shown. Gaps introduced to maximize alignment are indicated by dashes. The locations of the two phenylalanine pairs conserved in TBPs (solid arrowheads) are indicated. (B) Alignment of the C-terminal core domain of a selected number of TBPs with the corresponding domains of TBP-related factors. scTBP, S. cerevisiae TBP (accession no. P13393); dmTBP, D. melanogaster TBP (accession no. AAA68629); ceTBP, C. elegansTBP (accession no. AAA03582); dmTRF1, D. melanogaster TRF1 (accession no. Q27896); mmTRF2, Mus musculus TRF2 (accession no. AB017697); dmTRF2, D. melanogaster TRF2 (accession no. AF136569); ceTRF2, C. elegans TRF2 (accession no. AJ233443); tbTRF4, T. brucei TRF4 (GeneDB accession no. Tb10.61.0330). The locations of the two phenylalanine pairs conserved in TBPs (solid arrowheads), amino acids involved in DNA recognition (open squares), and TBP residues interacting with TFIIA (single line) and TFIIB (double line) are indicated. (C) Strict consensus of 278 most parsimonious trees from the analysis of a partial alignment of 127 parsimony informative characters. The number above the line represents the bootstrap resampling results using a heuristic parsimony analysis (percentage of 1,000 replicates). The number below the line is based on quartet puzzling steps (percentage of 10,000 replicates). Only values above 50% are shown. The analysis was done using PAUP 4.0b10.