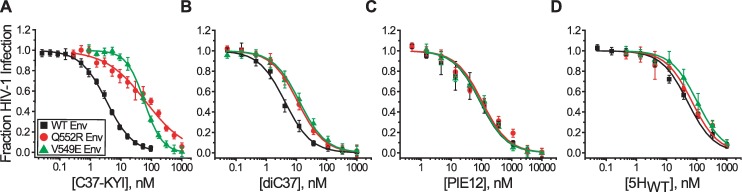
Fig 7. Effect of escape mutations Q552R and V549E on fusion inhibitor activity.
HIV-1 was pseudotyped with wild type EnvHXB2 (black) or Env variants containing the Q552R/N637K (red) or V549E/N637K (green) substitutions. Inhibitor titrations were performed with C37-KYI (A), di-C37 (B), PIE12 (C), and 5HWT (D) using HOS-CXCR4+ target cells. The regions of gp41 targeted by these inhibitors are shown in Fig 1C. Data points represent the mean ± SEM of three to seven independent experiments. In A, solid lines for wild type Env and the V549E mutant variant reflect fits of the data to a Hill equation; the red line for the Q552R mutant Env is a fit of the data to an inhibition model with both high and low affinity C37-KYI binding sites (see text). In B, C and D, the data have been fit to a simple Langmuir equation.