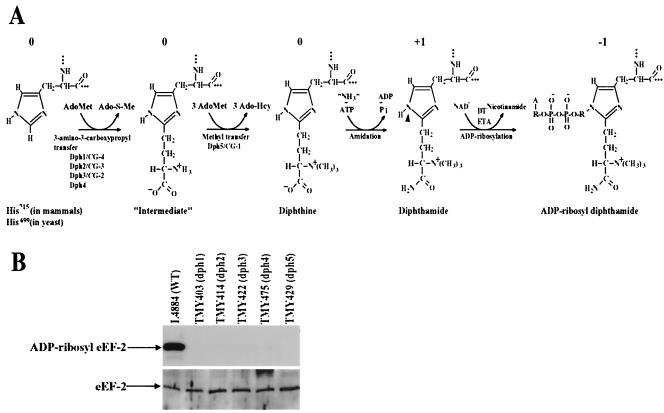
FIG. 1.
DPH1 to DPH5 genes are required for diphthamide biosynthesis. (A) Diphthamide biosynthesis and ADP ribosylation. N-1 (arrowhead) of the histidine imidazole ring of diphthamide is the site for ADP ribosylation by DT and ETA. Ado-S-Me, methylthioadenosine; Ado-Hcy, S-adenosylhomocysteine. In ADP-ribosyl diphthamide, A, adenine moiety; R, ribosyl moiety. The relative net charges of the modified His side chains are indicated above each species. (B) Precise deletions of each of DPH1 to DPH5 genes result in the yeast mutants being completely resistant to ADP ribosylation by DT. (Top) Yeast cell lysates were incubated with [adenylate-32P]NAD and DT at room temperature for 30 min. The reactions were terminated by boiling the lysates in SDS sample buffer, and the samples were analyzed by SDS-PAGE, followed by autoradiography. (Bottom) Deletions of each of the DPH1 to DPH5 genes do not affect overall expression of eEF-2 protein as evidenced by Western blot analysis using an anti-eEF-2 antibody (a gift of Angus Nairn, Rockefeller University).