Figure 1.
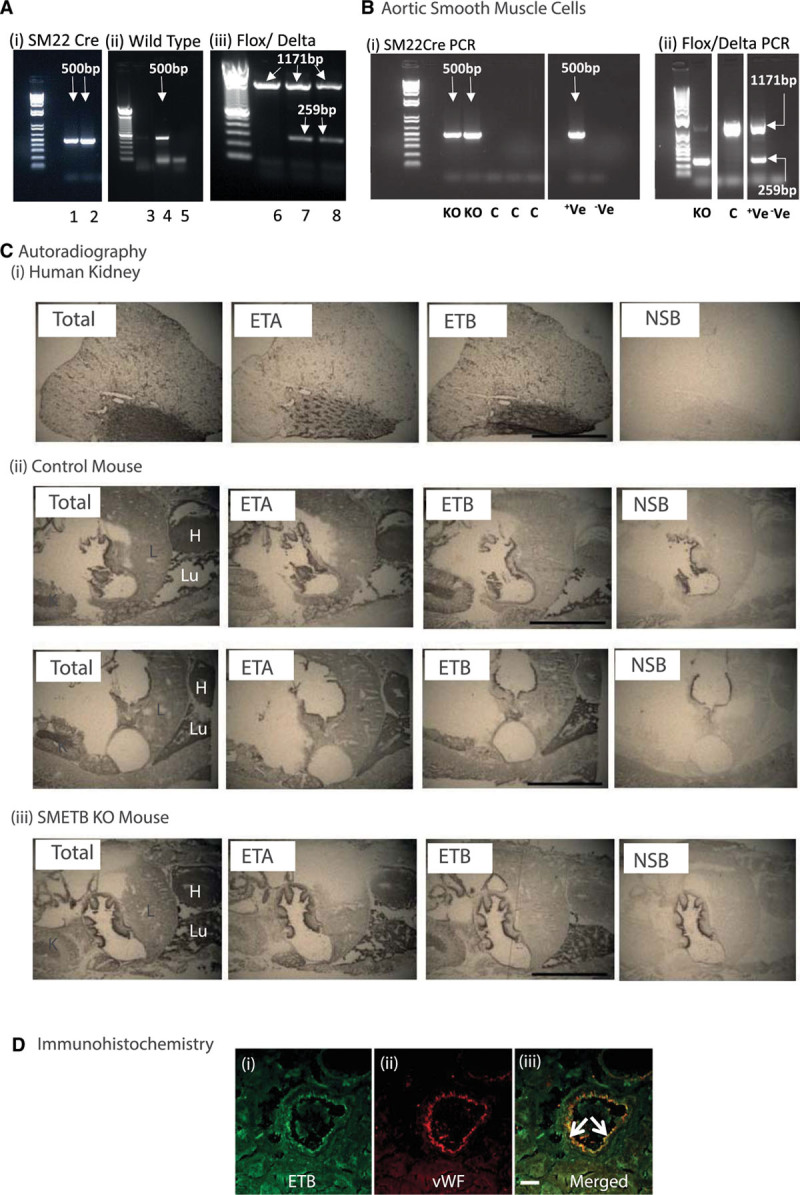
Selective endothelinB (ETB) receptor deletion from smooth muscle. A, Mice were genotyped for (i) SM22cre (band at 500 bp), (ii) wild-type (band a 500 bp), and (iii) flox (band at 1171 bp)/delta (band at 259 bp) alleles in ear clip DNA. (i) Samples 1 and 2 are cre-positive, (ii) sample 4 is positive for the wild-type allele; samples 3 and 5 are not, (iii) samples 7 and 8 are positive for both the flox and the delta band; sample 6 has only the flox band. B, Polymerase chain reaction (PCR) for cre and flox/delta bands in murine aortic smooth muscle cells isolated from smooth muscle ETB receptor knockout (SMETB KO) and control (C) mice. Control mice lacked cre and delta alleles, whereas SMETB KO expressed all 3. Standard DNA ladders have band sizes 1500–100 bp. C, Autoradiography showing maintained ETB ligand binding in SMETB KO lung and kidney (representative of n=3 mice/genotype). D, Confocal images of a coronary artery from an SMETB KO mouse stained for (i) ETB receptor (green) or (ii) the endothelial cell marker von Willebrand factor (vWF; red). Merged images (iii) show clear colocalization of ETB with the endothelium (arrows). There is no ETB staining in medial smooth muscle. Scale bar=50 µm. +Ve, positive control; –Ve, negative control; ETA indicates endothelinA; H, heart; K, kidney; L, liver; Lu, lung; and NSB, nonspecific binding.
