Fig. 2. RDG foci represent HJs in living cells.
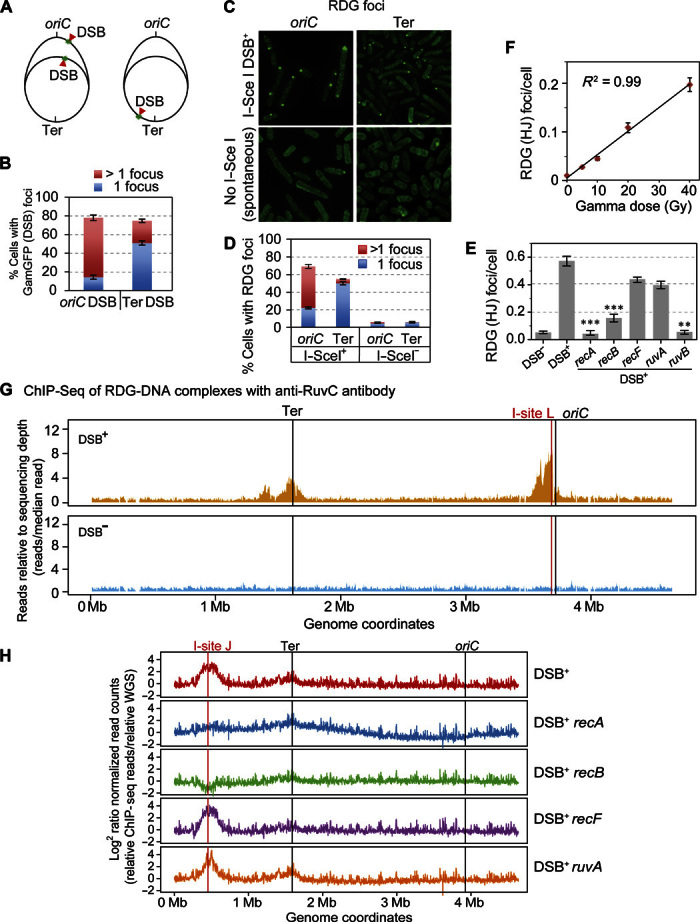
(A to E) Correlation of RDG foci with HR-DSB repair–generated HJs per living cell. (A) Strategy for E. coli chromosome cleavage with chromosomally encoded I–Sce I endonuclease at engineered cut sites (red arrows, DSB). Because of DNA replication, proliferating cells have more copies of oriC-proximal than oriC-distal DNA and thus will have more DSBs (and HR repair) per cell when cleaved by I–Sce I ori-proximally than ori-distally. (B) DSBs quantified as GamGFP focus, per Shee et al. (39), show that most of the cells with the oriC-proximal cut site have >1 GamGFP/DSB focus, and those with the ori-distal cut site have mostly 1 focus per cell, as previously described by Shee et al. (39). (C) Representative images of RDG foci after I–Sce I cleavage (top row) or spontaneous foci (bottom row). (D) I–Sce I–induced RDG foci are positively correlated with numbers of DSBs (and HJs) [quantification of images as in (B)], similarly to GamGFP foci in both oriC-proximal and oriC-distal sites. RDG foci increase with DSBs (P = 0.0001 for each locus, two-tailed unpaired t test), and more cells with >1 focus with ori-proximal than ori-distal cleavage (P = 0.0001, two-tailed unpaired t test). (E) HR protein dependence of I–Sce I/DSB–induced RDG foci. I–Sce I/DSB–induced RDG foci are reduced by a significant 26.1 ± 0.1 times in recA and 4.3 ± 0.1 (means ± SEM) times in recB null mutants indicating RecAB dependence, supporting their interpretation as HR-dependent foci formed during DSB repair. The dependence of RDG focus formation on RuvB supports their formation at four-way junctions. The independence of ruvA implies that RDG binds directly to DNA four-way junctions, as shown biochemically (44). **P < 0.01, ***P < 0.001, relative to DSB control, two-tailed unpaired t test. (F) RDG foci are positively correlated with dose of DSB-inducing γ radiation (R2 = 0.99; P < 0.001, Pearson’s correlation analysis). (G) RDG ChIP-seq shows that RDG localized DNA near a reparable I–Sce I–induced DSB (vertical red line) in the E. coli chromosome. I-site L, I–Sce I endonuclease site L. The large peaks disappear in DSB− cells (cells carrying the cut site but no I–Sce I enzyme). RDG ChIP-seq reads are normalized to the total reads in each sample and further normalized to the input genomic DNA. (H) RDG ChIP-seq shows enrichment at a different chromosomal I–Sce I cleavage site: I-site J, roughly halfway between the replication origin and terminus in the E. coli “right” replichore. The I–Sce I–induced RDG ChIP-seq peak is RecA- and RecB-dependent (indicating HR at a DSB), RecF-independent (indicating formation not at single-strand gaps), and RuvA-independent (supporting direct binding of RDG to four-way junctions, as for purified RuvC) (44). Figure S6B shows the DSB− (enzyme no cut site) controls. WGS, whole-genome sequencing.
