Figure 5.
Small-Angle X-Ray Scattering Analysis of the Myomesin:Obscurin-Like-1 Complex
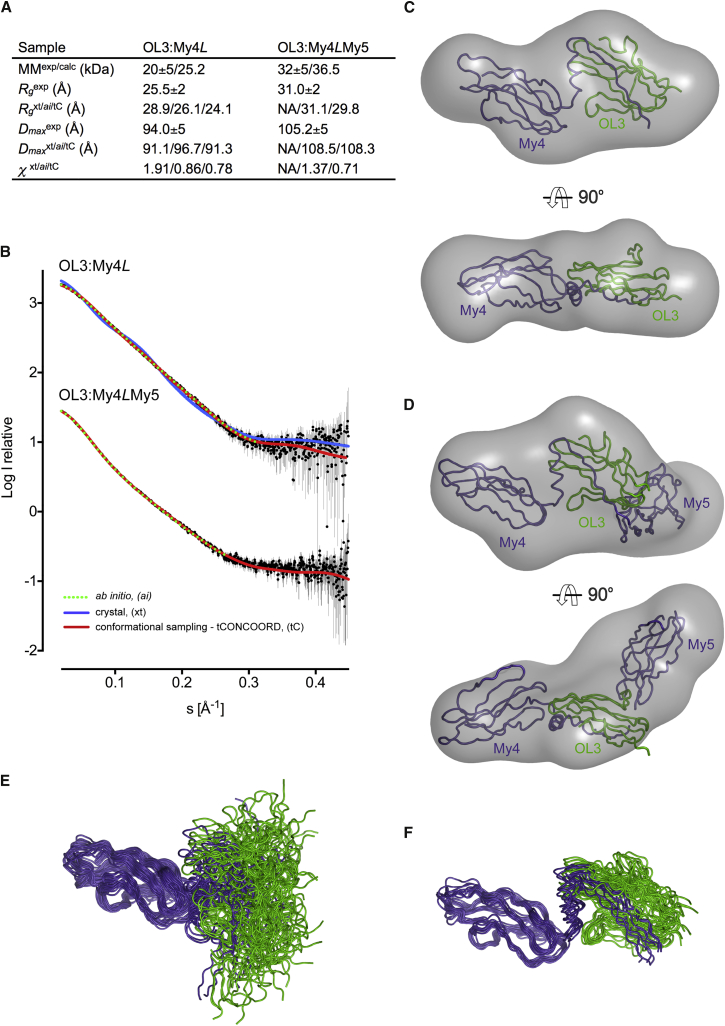
(A) Molecular parameters calculated from SAXS data. MM, Rg, Dmax are the molecular mass, radius of gyration, and maximum size, respectively. The superscript exp denotes experimental values while xt, ai, and tC refer to crystal, ab initio, and tCONCOORD fitted models, respectively. MMcalc is the theoretical MM computed from the protein sequence. χ is the discrepancy between experimental data and those computed from models.
(B) Experimental scattering intensities for OL3:My4L (upper curve) and OL3:My4LMy5 (lower curve) are displayed as dots with error bars. Curves computed from the crystallographic model and the best tC models are shown as solid blue and red lines, respectively, while the curve computed from the ab initio models is shown as a dotted green line.
(C) Cartoon-tube representation of a selection of 20 OL3:My4L tCONCOORD models (out of >30,000) aligned with respect to their My4 domain. My4L is in slate blue and OL3 is in green.
(D) Cartoon-tube representation of the ten OL3:My4L tCONCOORD models providing the best fit to the SAXS data (0.78 < χ < 0.81).
(E and F) OL3:My4L SAXS molecular envelope calculated from the ab initio model with a representative tCONCOORD model in two orthogonal orientations; (F) like (E) for OL3:My4LMy5.