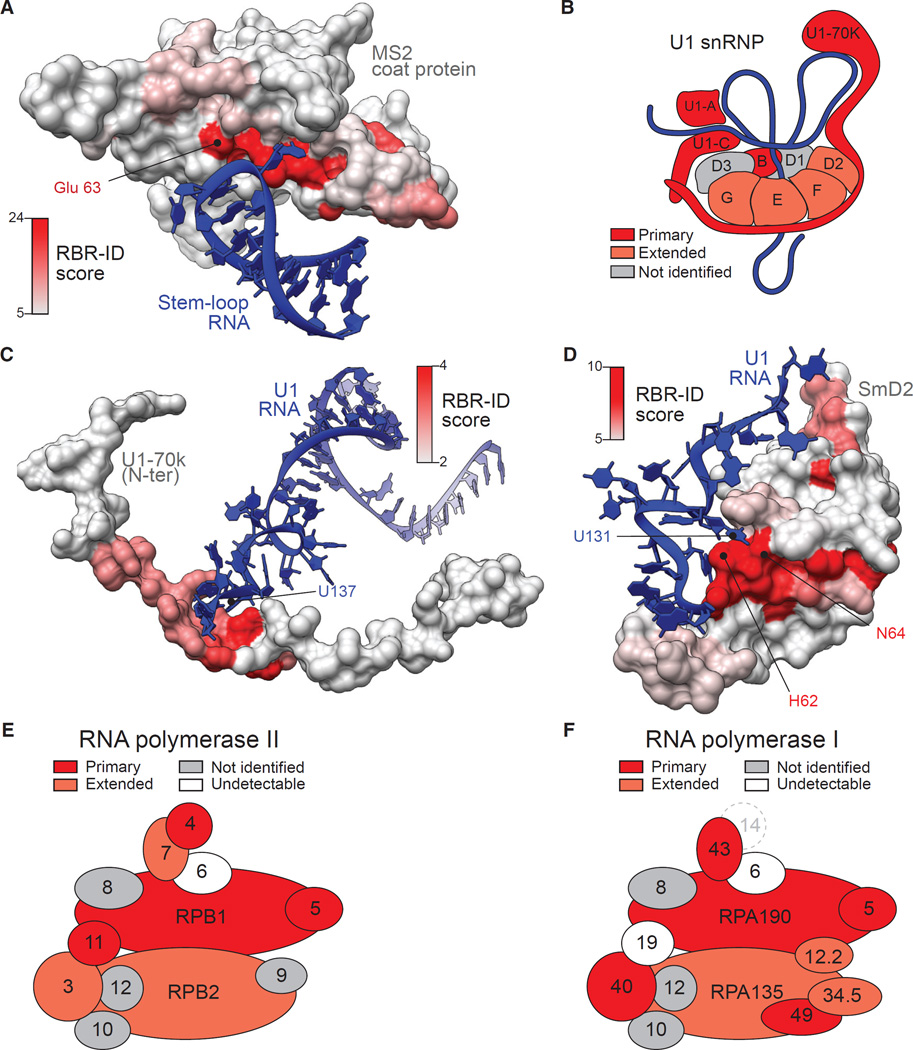
Figure 4. RBR-ID Maps the Sites of Protein-RNA Interactions In Vivo.
(A) The surface rendering of the MS2 coat protein in complex with its cognate RNA (PDB: 1ZDI; Valegârd et al., 1997) was color coded according to the residue-level RBR-ID score from the experiment shown in Figure 3.
(B) Schematic representation of the U1 snRNP particle (Kondo et al., 2015; Pomeranz Krummel et al., 2009). Subunits found in the primary list of RBR-ID candidates are in dark red; proteins in the extended list are in light red.
(C and D) Zoomed-in regions of the crystal structure of U1 snRNP (PDB: 4PJO; Kondo et al., 2015) showing protein surfaces color coded according to their RBR-ID score and interacting RNAs for two regions of U1-70K (C) and SmD2 (D).
(E and F) Schematic representation of the mammalian RNA pol II (E) and RNA pol I (F) complex according to Wild and Cramer (2012). Color coding is same as in (B). Subunits detected in the nuclear proteome, but not identified by RBR-ID, are in gray, undetectable subunits in white. The mammalian homolog for yeast RNA pol I subunit A14 (dashed circle) is unknown.
See also Figure S4.