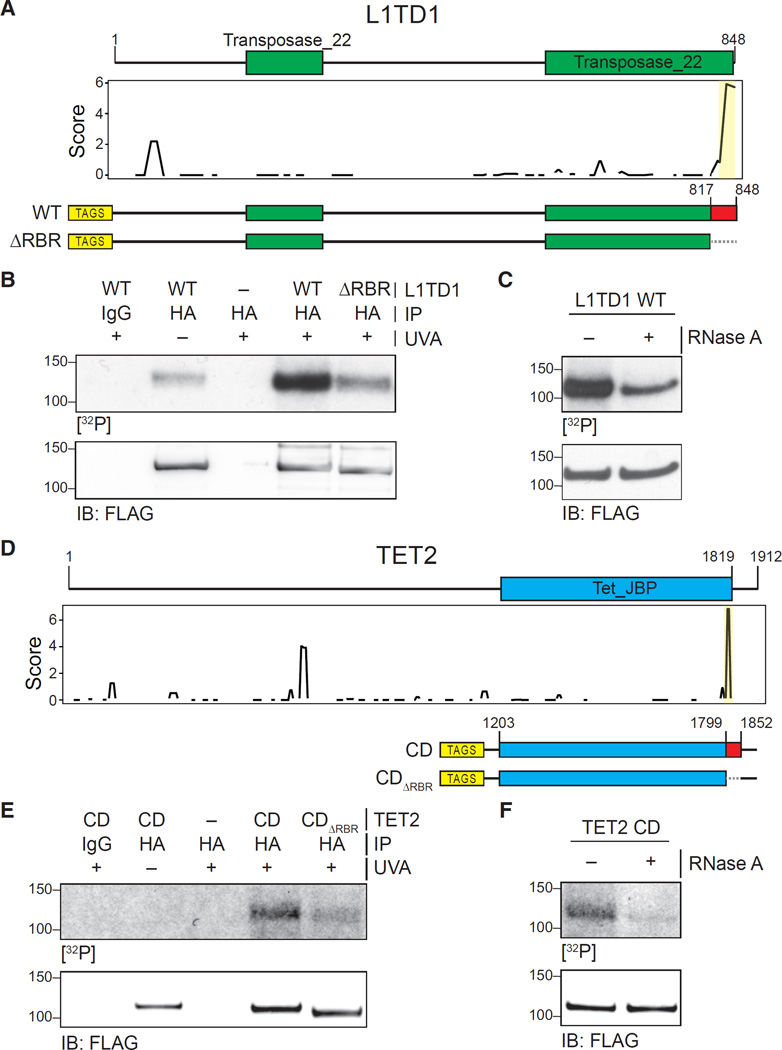
Figure 6. Validation of RBRs in L1TD1 and TET2.
(A) Primary sequence and known domains for L1TD1 (top); smoothed residue-level RBR-ID score plotted along the primary sequence (middle); and scheme of epitope-tagged WT and RBR-deleted (ΔRBR) constructs used for validation (bottom).
(B) PAR-CLIP of transiently expressed WT and ΔRBR L1TD1 in HEK293 cells. Autoradiography for 32P-labeled RNA (top) and control western blot (bottom).
(C) PAR-CLIP for WT L1TD1 with and without treatment with RNase A (top) and control western blot (bottom).
(D) Primary sequence and known domains for TET2 (top); smoothed residue-level RBR-ID score plotted along the primary sequence (middle); and scheme of epitope-tagged catalytic domain fragment (CD) and RBR-deleted (CDΔRBR) constructs used for validation (bottom).
(E) PAR-CLIP of transiently expressed TET2-CD and TET2-CDΔRBR in HEK293 cells. Autoradiography for 32P-labeled RNA (top) and control western blot (bottom).
(F) PAR-CLIP for TET2 CD with and without treatment with RNase A (top) and control western blot (bottom).