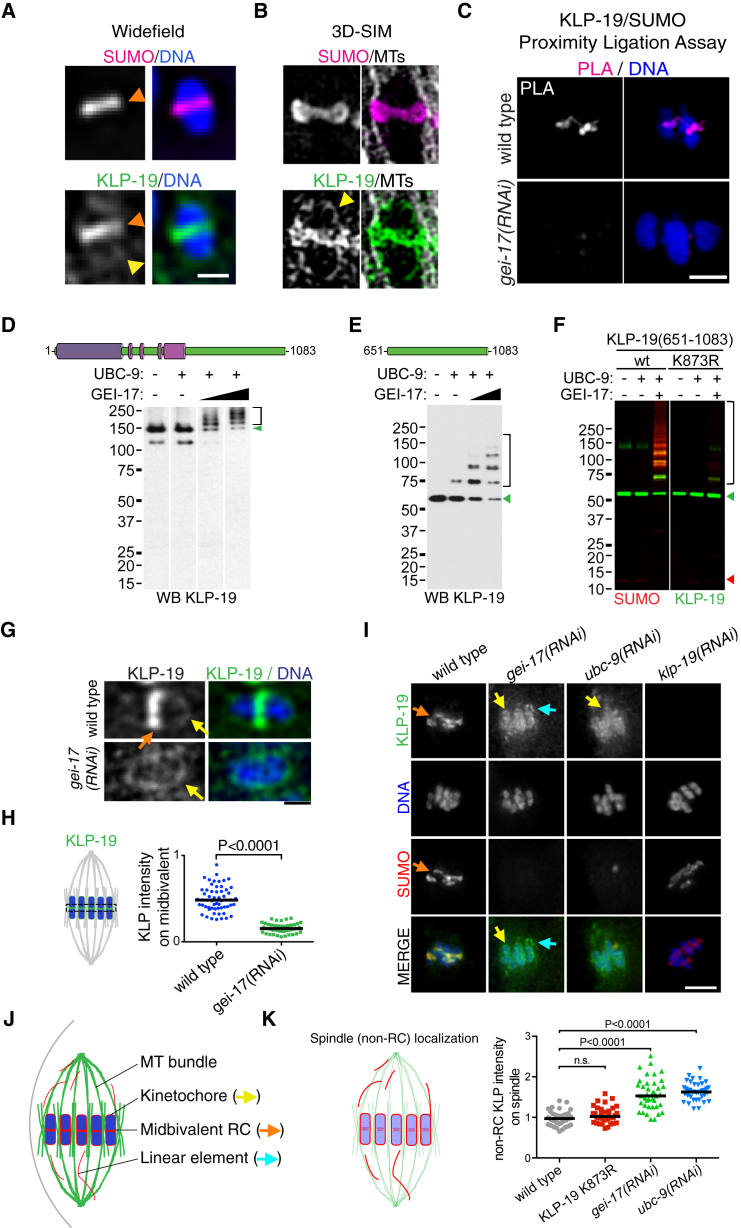
Figure 3.
KLP-19 Is a SUMO Substrate and Its Ring Localization Depends on SUMO Conjugation
(A) Widefield images of a single bivalent stained for SUMO (top) and KLP-19 (bottom). The yellow arrow points to the kinetochore, while the orange arrow marks the midbivalent. The scale bar represents 1 μm.
(B) 3D-SIM images of a single bivalent stained for SUMO (magenta) or KLP-19 (green) and α-tubulin (gray).
(C) Proximity ligation assay of the KLP-19-SUMO interaction in control and gei-17(RNAi) worms. The bivalents during the first meiotic division are shown. The scale bar represents 2 μm.
(D) The cartoon depicts the domain architecture of KLP-19. The motor domain is shown in purple and the putative coiled-coil domains in pink. Full-length, recombinant KLP-19 was incubated in the presence of SUMO (20 μM), E1 enzyme (100 nM), UBC-9 (140 nM), and GEI-17 (12.5 and 25 nM). An aliquot of the reaction was run on an SDS-PAGE, and western blot was performed with an anti-KLP-19 antibody. The green arrowhead points to unmodified KLP-19, and the square bracket denotes SUMO-modified KLP-19. See also Figures S4A, S4B, S4D, and S4E.
(E) In vitro SUMO conjugation reactions were performed with KLP-19(651–1,083) as substrate. The reactions were incubated in the presence of SUMO (20 μM), E1 enzyme (100 ng), UBC-9 (140 nM), and increasing amounts of GEI-17 (12.5 and 25 mM). The reactions were developed as in (A). The green arrowhead points to unmodified KLP-19, and the square bracket denotes SUMO-modified KLP-19. See also Figures S4A, S4C, S4D, and S4F.
(F) Wild-type and K873R KLP-19(651–1,083) were subject to GEI-17-dependent in vitro SUMO conjugation, using the same conditions as above and 12.5 mM GEI-17. The reactions were then run on a gel, transferred to a membrane, and developed by two-color near-infrared western blotting (LICOR). KLP-19 is shown in green and SUMO in red. The red arrowhead indicates free SUMO; the green arrowhead points to unmodified KLP-19; and the square bracket denotes SUMO-modified KLP-19. See also Figures S3A–S3I, S4G, and S4H.
(G) KLP-19 localization in the midbivalent is dependent on GEI-17. Metaphase I-arrested oocytes were stained for KLP-19, SUMO, and DNA as indicated. The yellow arrow points to the kinetochore, while the orange arrow marks the midbivalent.
(H) Quantitation of the KLP-19 intensity in the midbivalent was analyzed using the Mann-Whitney test. The black lines indicate the median.
(I) KLP-19 localization was analyzed after GEI-17 or UBC-9 depletion. An orange arrow signals KLP-19 on the midbivalent. The yellow arrow indicates kinetochore localization. The blue arrow indicates the linear elements (see main text for details). The scale bar represents 4 μm. See also Figure S5D.
(J) Schematic depicting the procedure for quantifying KLP-19 residing outside the RC (“non-RC”). The microtubule bundles, kinetochores, midbivalents, and linear elements are shown next to the colored arrows used throughout the figure to highlight them. The whole spindle area was selected using α-tubulin as a guide, and the midbivalents were selected from the DNA channel. After substracting the background-corrected midbivalent signal to the background-corrected spindle signal, we obtained the non-RC KLP-19 intensity.
(K) KLP-19 re-localizes to kinetochores and linear elements after UBC-9 or GEI-17 depletion. The data for non-RC KLP-19 intensity are shown as a dot plot, and the samples were compared using a Kruskal-Wallis test, followed by Dunn’s post-test. The black lines denote the medians.