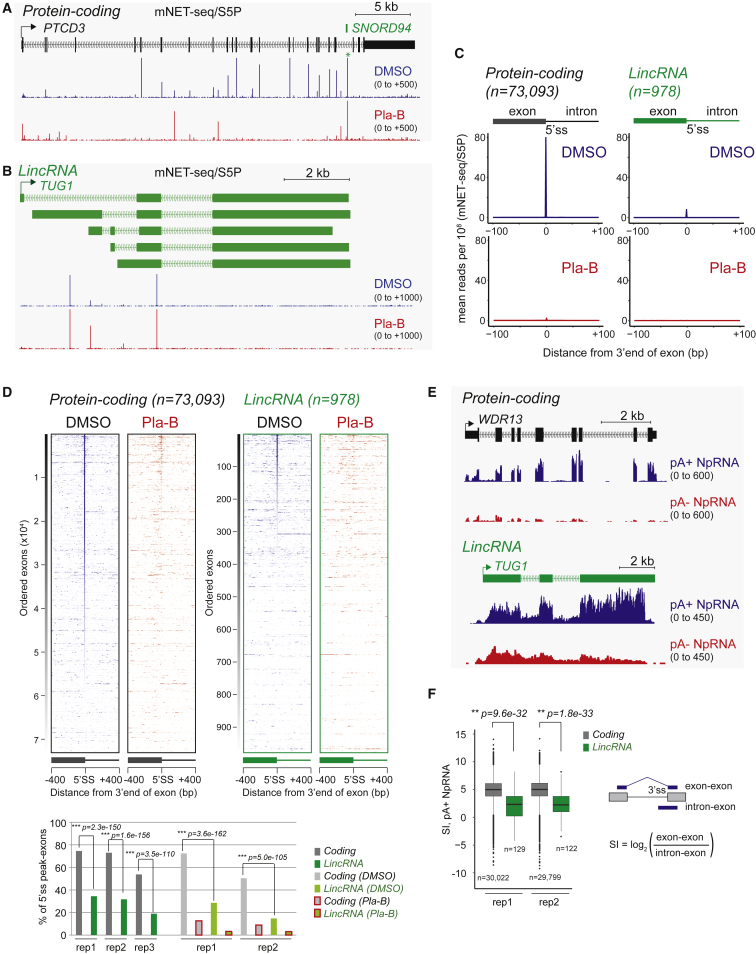
Figure 2.
lincRNAs Are Inefficiently Spliced
(A and B) (A) mNET-seq/S5P analysis of protein-coding gene PTCD3 and (B) lincRNA TUG1. HeLa cells were treated with Pla-B (red) or DMSO control (blue). Only sense transcripts are shown.
(C) Meta-analysis across exon-intron junctions (5′ss) of annotated introns for protein-coding TUs versus lincRNAs.
(D) Heatmaps for protein-coding versus lincRNA genes aligned to 5′ss −400 to +400 nt upstream and downstream. Percent of introns showing co-transcriptional 5′ss peaks is shown below, including all data repetitions, either with untreated, DMSO mock-treated, or Pla-B-treated HeLa cells.
(E) pA+ and pA− NpRNA-seq profiles are shown for WDR13 versus lincRNA TUG1.
(F) Splicing index from pA+ NpRNA-seq for protein-coding and lincRNA TUs (duplicates shown).
See also Figure S2.