Fig. 2.
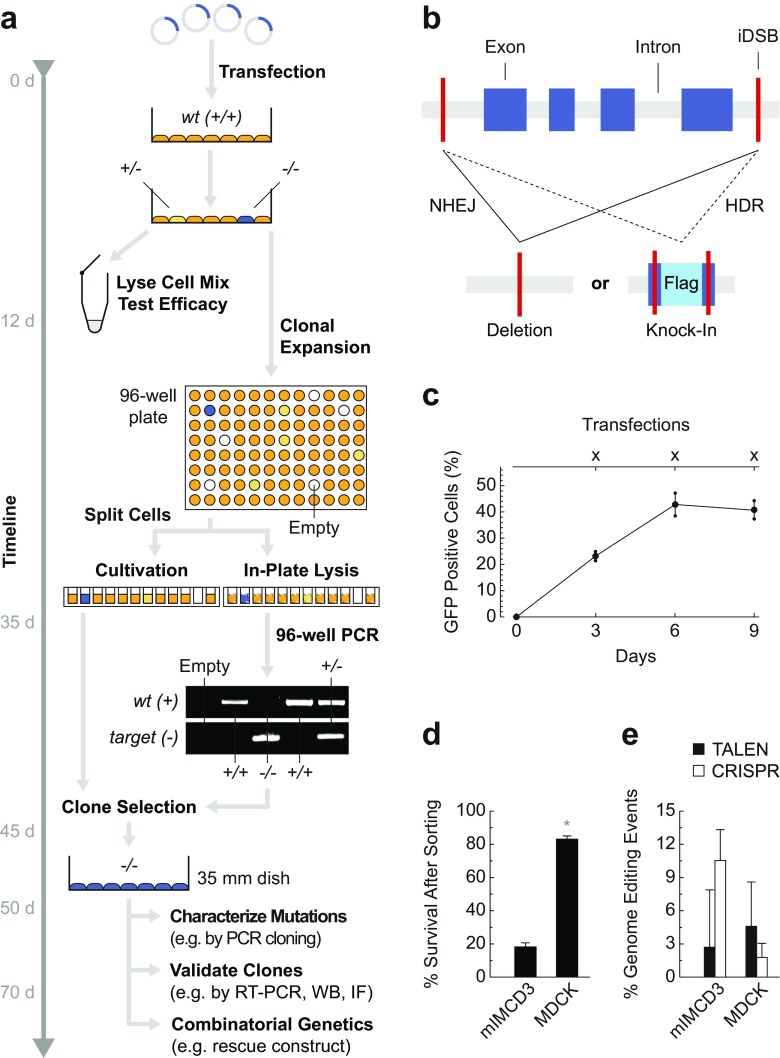
Genome editing of MDCK and mIMCD3 renal epithelial cells. a Here, we provide a practical guide to generate genetically modified renal epithelial cell culture models. Genome editing requires transfection of programmable sequence-specific nucleases to induce DNA double strand breaks (iDSB) [15, 21]. After PCR-based testing for genome editing efficacy, cells are singularized in 96-well plates and clones are PCR-screened for targeted alleles. Targeted clones (−/−) are selected and validated by independent methods. d days, WB Western blot, IF immunofluorescence. See Supplementary Methods for a step-by-step protocol. b Nuclease-induced DSBs are repaired by non-homologous end joining (NHEJ) or homology directed repair (HDR). Both mechanisms can be exploited for genome editing [15, 21]. For example, if two DSBs are induced, error prone NHEJ may skip the middle piece causing a deletion; or, if two DSBs are induced and a targeting vector is provided, precise HDR may incorporate exogenous sequence elements (here, a Flag epitope tag for biochemical protein detection and isolation) into the genome. c Repeated transfection of cells increases the proportion of transfected cells significantly. MDCK cells were repeatedly nucleofected with GFP and fluorescent cells were counted using an automated cell counter (N = 3; for each N we analyzed ≥2′000 cells). d In contrast to MDCK cells, mIMCD3 cells are more sensitive to the transfection and sorting procedure requiring higher cell numbers a priori (N = 5; wells evaluated after single cell sorting / N ≥ 1′000). e To evaluate genome editing efficacy in MDCK and mIMCD3 cells we targeted three genes per cell line with both, TALEN and CRISPR technology. Target alleles for TALENs and CRISPR were similar, but not identical, due to different genomic binding site requirements. We aimed for large deletions (≥10 kb) or HDR-mediated integrations of targeting vectors. Correctly targeted heterozygous or homozygous clones were counted. Minor NHEJ-mediated sequence changes were not considered. Significant genome editing events were observed in 3.19% of MDCK and 6.62% of mIMCD3 cells (N = 3 per cell line and genome editing technology; for each N we analyzed between 90 and 1′101 clones). Data are presented as means with s.e.m.