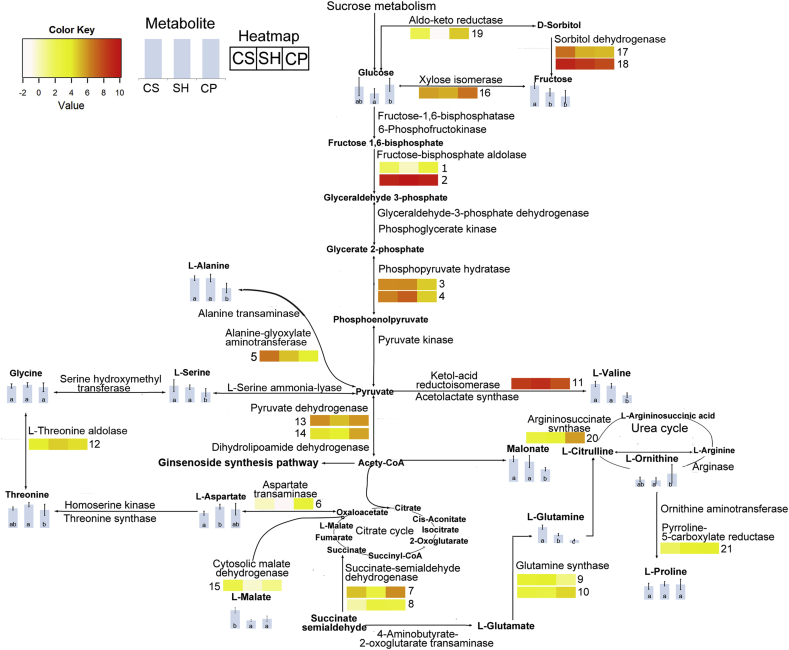
Fig. 4.
Comparative analysis of primary metabolites and genes involved in their biosynthesis pathways in three ginseng cultivars, Cheongsun (CS), Sunhyang (SH), and Chunpoong (CP). The contents in adventitious roots from three ginseng cultivars of compounds detected by GC-MS analysis are represented in vertical bar graphs. Genes found in the CP root transcriptome are shown on the map. Genes with significantly different expression levels among the three cultivars are represented in the heatmaps. The expression levels of unigenes putatively encoding the enzymes were determined using FPKM values, and the mean FPKM values of significantly differently expressed genes (p < 0.05) were transformed using log2 and represented on the heatmaps using a white-to-red gradient. The numbering in the heatmap follows the system used in Table 5. FPKM, fragments per kilobase of exon per million fragments; GC-MS, gas chromatography–mass spectrometry.