Figure 2.
EBF3 p.Arg163Gln and p.Arg163Leu Impair Transcriptional Activation
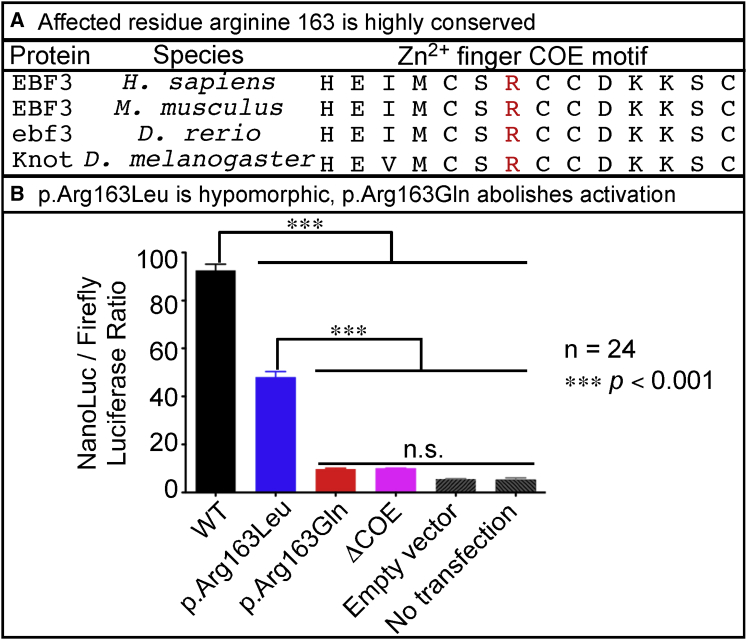
(A) Affected residue Arg163 in the Zn2+ finger COE motif is highly conserved across vertebrates and invertebrates.
(B) Activation of reporter-gene expression in HEK293 cells was assessed as the ratio of NanoLuc to firefly luciferase according to the Promega NanoGlo Dual Reporter protocol and was measured on the TD 20/20 Luminometer. The cDNAs encoding WT EBF3 and the EBF3 p.Arg163Gln and p.Arg163Leu variants had a stop codon to ensure that the proteins were untagged to minimize off-target effects from a protein tag. The cDNAs were subcloned into the mammalian expression vector, pcDNA-DEST40. Two synthetic oligonucleotides containing the imperfect palindromic COE binding sequence were used to generate six concatamerized COE binding sites in the NanoLuc vector, pNL3.1. The pGL4.53 firefly luciferase vector was used as an internal transfection control. Additional experimental controls included EBF3 with deletion of the Zn2+ finger COE motif (denoted as ΔCOE). As transfection background controls, we either transfected only the pNL3.1 with six COE binding sites and pGL4.53 but no cDNA expression vectors (denoted as “empty vector”) or did not transfect any vectors (denoted as “no transfection”). A 92-fold induction was observed with WT EBF3 (black). However, EBF3 p.Arg163Leu caused only a 45-fold induction (blue), indicating a partial loss of transcriptional activation. EBF3 p.Arg163Gln showed a very poor induction of transcription (red) similar in level to the transfection background (gray) and that of EBF3 ΔCOE (magenta) controls, indicating severe loss of activity. Data represent the mean ± SEM. n = 24 (six replicates per four separate transfections per experimental condition); ∗∗∗p < 0.001 via one-way ANOVA with Tukey’s post hoc analysis; n.s., no significant difference.