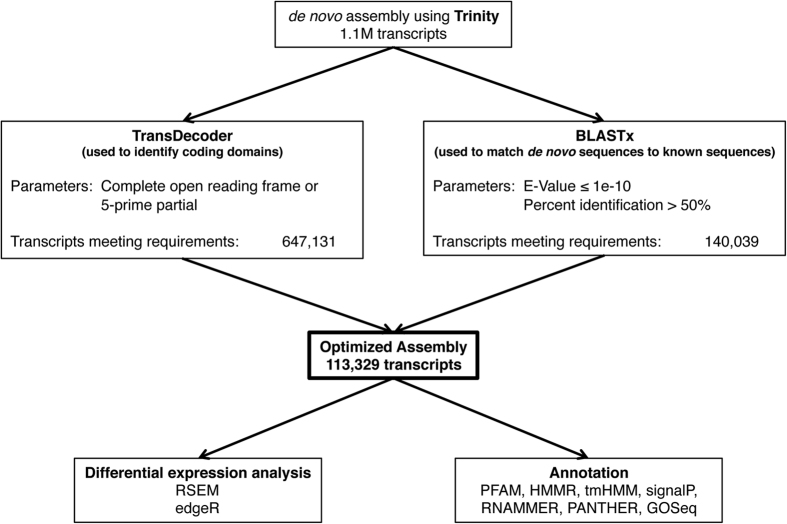
Figure 1. Schematic of de novo assembly optimization and analysis.
After initial de novo assembly using Trinity, we optimized the assembly using several programs to omit falsely assembled sequences or sequences that were not likely to code for an actual gene. After optimization, we used RSEM to generate expected counts of each transcript from the raw reads and used those reads to calculate differential expression between males and females using edgeR. Annotation of the optimized assembly was completed using a series of annotation steps, PANTHER, and GOSeq.