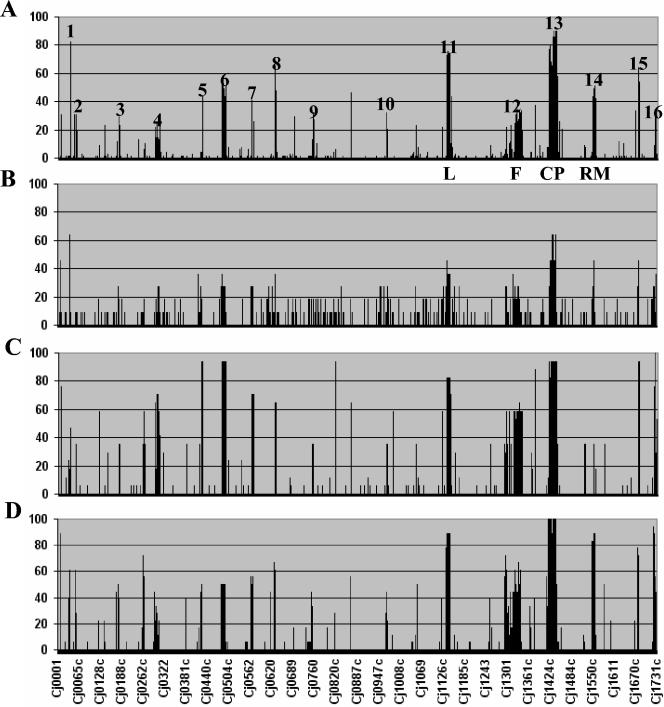
FIG. 2.
Survey of genetic variability in C. jejuni. Divergent genes were determined for each strain, and the percentage of strains showing divergence at each gene position was calculated and plotted as a histogram according to their position on the genome strain NCTC 11168 (18). The results for data set III (A), data set I (7) (B), data set II (14) (C), and data set IV (22) (D) are shown. Most variable genes map to 16 hypervariable regions in the C. jejuni NCTC 11168 genome (Table 2). These include the lipooligosaccharide biosynthesis locus (L), the flagellar biosynthesis locus (F), the capsular polysaccharide biosynthesis locus (CP), and the restriction-modification locus (RM). Although similar trends were observed for all four data sets, data set III (A) shows better resolution because of the larger sample size.