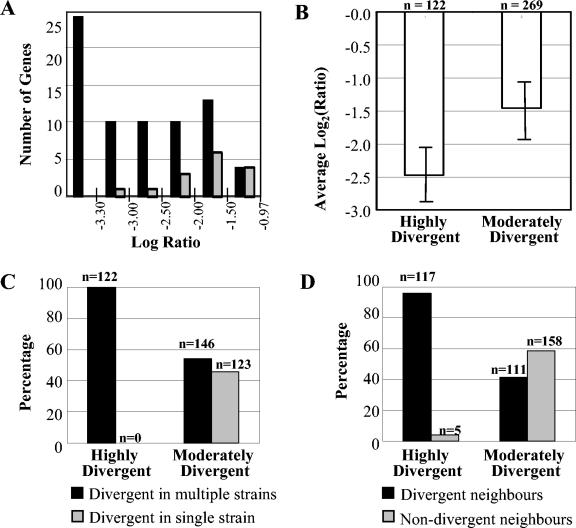
FIG. 4.
Association between high levels of divergence, the occurrence of codivergent clusters, and divergence in multiple strains. (A) Genes which were divergent in experiments with strain RM1221 and NCTC 11168 CGH (log ratio < −0.97) were binned by log ratio value. The BLAST server at TIGR was used to examine the homology of the corresponding NCTC 11168 genes to the RM1221 genome sequence. Gray bars, the number of genes in each bin for which BLAST hits indicated detectable sequence identities; black bars, genes without BLAST hits in the RM1221 genome. On the basis of these data, a log ratio <−3.3 was used as a cutoff to define HD or absent genes from CGH data. (B) Average log ratios [log2(test signal/control signal)] for HD and MD genes. A statistically significant difference in the average log ratios for the two groups can be observed. (C) Percentage of divergent genes that were variable in multiple strains. HD genes are exclusively found to be divergent in multiple strains (100%; 122 of 122). In contrast, MD genes have a similar likelihood of being divergent in a single strain or in multiple strains (45.7 and 54.3%, respectively). (D) Percentage of divergent genes that were adjacent in the C. jejuni NCTC 11168 genome. The majority of HD genes have codivergent neighbors (95.9%; 117 of 122), whereas this value is only 41.3% among MD genes. The figure is based on raw microarray CGH data for data sets II and III.