Abstract
Different typing schemes for Campylobacter spp. were evaluated with 70 outbreak and sporadic isolates. The discriminatory indexes were 0.944 (by pulsed-field gel electrophoresis), 0.920 (by genotyping of the flagellin A gene), 0.902 (by genotyping of flaB), and 0.886 (by multilocus sequence typing). Cross-classification gave 94.77 or 95.82% (PFGE-flaA or PFGE-flaB) concordance. flaA was overdiscriminatory in three cases, most probably due to intragenomic recombination.
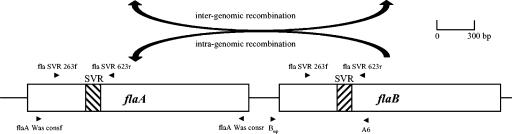
To differentiate sporadic from epidemiologically related Campylobacter infections, a rapid and discriminatory typing method is required to identify sources of human infection and to determine the routes of infection. Pulsed-field gel electrophoresis (PFGE) has proven to be useful and discriminatory and is therefore considered the present gold standard (17). However, PFGE is laborious, and even with standardized protocols the interlaboratory comparison of PFGE remains difficult. Methods based on DNA sequencing are fast, eliminate experimental variation, and facilitate interlaboratory comparisons (5). Recently, it was shown that the genotyping of the short variable region (SVR) of the flagellin A gene (flaA) provides adequate discrimination in short-term epidemiology (7). However, its use is questioned because of the known intra- and intergenomic recombination within the flagellin genes (1, 6, 16). The flagellum is encoded by two highly homologous genes (flaA and flaB) (Fig. 1) of approximately 1,730 bp joined by an intervening segment of approximately 200 bp (6). The nucleotide sequence of a 321-bp region of the flaA and flaB genes was determined for each isolate. This sequence encompassed the SVR extending from fla nucleotide positions 283 to 603 (inclusive) (3). The flagellin B gene (flaB) is, in contrast to flaA, not essential for motility and associated pathogenicity and is thought to be a genetic reservoir for flaA (1). Therefore, it can be assumed that flaB is a more stable marker. The aim of this study was to evaluate the potential of flaB typing in comparison to that of flaA typing.
FIG. 1.
Scheme of the genetic organization of the flagellin genes showing locations of primers used for PCR amplification and sequencing.
(This study was presented in part at the 104th General Meeting of the American Society for Microbiology, New Orleans, La., 23 to 27 May 2004.)
Partial DNA sequences (SVR, 321 bp) of flaA and flaB from 36 Campylobacter isolates (including 3 controls) from three documented outbreaks (Germany in 1997 and 2000 [14, 15] and Kansas in 1988 [12]) were analyzed. The control isolates were matched to each outbreak setting. Also, 34 consecutive strains from 13 sporadic cases of campylobacteriosis isolated in 2002 and 2003 were examined to assess the target's genetic stability. The flaA and flaB sequencing was essentially carried out as previously described (6, 17). Briefly, for amplification of flaA the consensus primers described by Wassenaar et al. (17) were used, and for flaB the primers Bup and A6 were used. For sequencing either the forward primer fla SVR 263f (5′-AAR GCT ATG GAT GAG CAA YTW AAA AT-3′) or the reverse primer fla SVR 623r (5′-CCA AGW CCT GTT CCW ACT GAA G -3′) were applied for both genes (Fig. 1). The alleles were assigned using the Campylobacter FlaA Variable Region Database (http://phoenix.medawar.ox.ac.uk/flaA/). In addition, the isolates were characterized by PFGE and by multilocus sequence typing (MLST). PFGE was performed according to the electrophoretic conditions previously described by Ribot and colleagues with SmaI as the restriction enzyme (10). Isolates differing at one or more bands were considered to be different. The macrorestriction patterns were arbitrarily designated with capital letters. For MLST, sequencing of seven housekeeping genes (aspA, glnA, gltA, glyA, pgm, tkt, and uncA) and assignment of sequence types (ST) were carried out in agreement with the protocol of Dingle et al. (2). The typing results of 42 C. jejuni isolates for which there was complete data (PFGE, MLST, flaA, and flaB typing results) were compared by assessing the discriminatory index (DI) (4) and the cross-classification results (11).
An overview of the typing results is shown in Table 1, which also includes the epidemiologic information of all Campylobacter isolates analyzed in this study. Mixed infections with two different C. jejuni strains occurred in sporadic cases 8 and 11. In sporadic case 12 even a coinfection with two Campylobacter species was observed. These findings are consistent with the observation that potential sources of Campylobacter infection may be contaminated with more than one strain (9).
TABLE 1.
Comparison of the PFGE, MLST, flaA, and flaB sequencing results for sporadic and outbreak-associated Campylobacter isolatesi
| Species and strain no. | Origin | Epidemiologic implication | PFGE (SmaI)a | MLST STb | flaA allelec | flaB allelec | Comment |
|---|---|---|---|---|---|---|---|
| C. jejuni | |||||||
| B01751 | Human | Y | S | ST 21 | 245 | 226 | Outbreak 1d |
| B01755 | Human | Y | S | ST 21 | 245 | 226 | Outbreak 1 |
| B01758 | Human | Y | S | ST 21 | 226 | 226 | Outbreak 1 |
| B01759 | Human | Y | S | ST 21 | 226 | 226 | Outbreak 1 |
| B01766 | Cow feces | Y | S | ST 21 | 245 | 226 | Outbreak 1 |
| B01768 | Cow feces | Y | S | ST 21 | 245 | 226 | Outbreak 1 |
| B01769 | Cow feces | Y | S | ST 21 | 245 | 226 | Outbreak 1 |
| B01770 | Cow feces | Y | S | ST 21 | 245 | 226 | Outbreak 1 |
| B01513 | Human | N | T | ST 823 | 117 | 117 | Outbreak 1 (control) |
| B02098 | Human | Y | A | ST 575 | 239 | 177 | Outbreak 2e |
| B02100 | Human | Y | A | ST 575 | 239 | 177 | Outbreak 2 |
| B02122 | Skim cream | N | L | ST 861 | 121 | 103 | Outbreak 2 (control) |
| B02184 | Cow feces | N | E | ST 45 | 2 | 307 | Outbreak 2 (control) |
| *D2678 | Human | Y | 8f | ST 21 | 49 | 326 | Outbreak 3 |
| *D2692 | Raw milk | Y | 8 | ST 21 | 326 | 326 | Outbreak 3 |
| Ob5647 | Human | Y | V | ST 824 | 105 | 260 | Sporadic 1 |
| Ob5815 | Human | Y | V | ST 824 | 105 | 260 | Sporadic 1 |
| Ob6055 | Human | Y | V | ST 824 | 105 | 260 | Sporadic 1 |
| Ob6965 | Human | Y | W | ST 21 | 32 | 103 | Sporadic 2 |
| Ob7119 | Human | Y | W | ST 21 | 32 | 103 | Sporadic 2 |
| Ob7420 | Human | Y | W | ST 21 | 32 | 103 | Sporadic 2 |
| EB1410 | Human | Y | M | ST 822 | 5 | 5 | Sporadic 3 |
| EB1430 | Human | Y | M | ST 822 | 5 | 5 | Sporadic 3 |
| EB1599 | Human | Y | M | ST 822 | 5 | 5 | Sporadic 3 |
| A57511 | Human | Y | N | ST 658 | 5 | 5 | Sporadic 4 |
| A58515 | Human | Y | N | ST 658 | 5 | 5 | Sporadic 4 |
| SL5512 | Human | Y | O | ST 50 | 36 | 36 | Sporadic 5 |
| SL5546 | Human | Y | O | ST 50 | 36 | 36 | Sporadic 5 |
| EB87 | Human | Y | P | ST 572 | 14 | 96 | Sporadic 6 |
| EB101c | Human | Y | Q | ST 572 | 14 | 96 | Sporadic 6 |
| EB101j | Human | Y | Q | ST 572 | 14 | 96 | Sporadic 6 |
| A1689 | Human | Y | R | ST 607 | 14 | 14 | Sporadic 7 |
| A2891 | Human | Y | R | ST 607 | 14 | 14 | Sporadic 7 |
| Ob4699 | Human | Y | AA | ST 21 | 121 | 103 | Sporadic 8 (mixed infection) |
| Ob5876 | Human | Y | Y | ST 50 | 36 | 36 | Sporadic 8 |
| Ob6054 | Human | Y | Y | ST 50 | 36 | 36 | Sporadic 8 |
| Ob6560 | Human | Y | Y | ST 50 | 36 | 36 | Sporadic 8 |
| Ob6343 | Human | Y | Z | ST 22 | 232 | 309 | Sporadic 9 |
| Ob6563 | Human | Y | Z | ST 22 | 232 | 309 | Sporadic 9 |
| Ob4819 | Human | Y | F | ST 52 | 57 | 57 | Sporadic 10 |
| *Ob5646 | Human | Y | —g | ST 52 | 57 | 57 | Sporadic 10 |
| A21989 | Human | Y | J | ST 658 | 5 | 5 | Sporadic 11 (mixed infection) |
| A23530 | Human | Y | I | ST 572 | 105 | 260 | Sporadic 11 |
| A25293 | Human | Y | I | ST 572 | 105 | 260 | Sporadic 11 |
| Ob6185 | Human | Y | L | ST 842 | 121 | 103 | Sporadic 12 (mixed infection) |
| C. coli | |||||||
| *Ob6057 | Human | Y | G | NTh | 228 | 308 | Sporadic 12 |
| *Ob6562 | Human | Y | X | NT | 229 | 310 | Sporadic 13 |
| *Ob7415 | Human | Y | X | NT | 229 | 310 | Sporadic 13 |
| *Ob7852 | Human | Y | X | NT | 229 | 310 | Sporadic 13 |
PFGE type names are arbitrary.
MLST sequence types were assigned using the MLST website (http://www.mlst.net).
fla alleles were assigned using the Campylobacter FlaA Variable Region Database (http://phoenix.medawar.ox.ac.uk/flaA/).
Three further outbreak isolates with epidemiological implications (Y) showed the same typing results (MLST was not done).
Eighteen further outbreak isolates with epidemiological implications (Y) showed the same typing results (MLST was not done).
PFGE types were numbered arbitrarily (relation to the other PFGE patterns is unknown).
Did not restrict with SmaI.
NT, not typeable.
Strains marked by an asterisk were not included for discriminatory index and concordance calculations. Boldface indicates intragenomic recombination events.
Using the primers FLA4F and FLA625RU originally proposed by Meinersmann et al. for amplification of the flaA SVR frequently resulted in sequences with ambiguous bases (7), most probably due to flagellin gene paralogs (8). Changing to the recently published consensus flaA forward and reverse primers gave better results (17). No single ambiguous base was observed when using the Bup and A6 primers for flaB amplification (6). One isolate was resistant to SmaI digestion and therefore was not typeable by PFGE. For the other 42 C. jejuni isolates, PFGE was most (DI, 0.944) and MLST was least discriminatory (DI, 0.886). flaA and flaB typing gave intermediate DI results of 0.920 and 0.902, respectively (Table 2). The DI depends on the number of types and on the homogeneity of frequency distribution of strains into types (13). Therefore, although MLST gave more types than fla typing, the DI was lower because of the inhomogeneous distribution of the STs. Ideally, the DI should be calculated using a test population that includes epidemiologically unrelated strains (13). Obviously, this is not true in our study; therefore, the absolute DI figures should be treated with caution. Nevertheless, the relative ordering of the typing schemes according to the DIs is meaningful.
TABLE 2.
Resolving power of PFGE, MLST, flaA, and flaB typing schemes for C. jejuni (n = 42)
| Method | No. of types | No. of most frequent type | Index of diversity (95% CIa) |
|---|---|---|---|
| PFGE | 18 | 8 | 0.944 (0.909-0.979) |
| MLST | 14 | 12 | 0.886 (0.825-0.948) |
| flaA | 13 | 6 | 0.920 (0.895-0.944) |
| flaB | 12 | 8 | 0.902 (0.871-0.934) |
CI, confidence interval.
The cross-classification of all possible pairs of PFGE and flaA or flaB gave 94.77 and 95.82% concordance. PFGE and MLST results were only 93.96% concordant (Table 3). In two outbreaks, flaA was overdiscriminatory compared to the other employed typing schemes, whereas flaB gave correct typing results. In outbreak 1, two isolates (B01766 and B01768; isolated from independent sources) were different from the others by flaA SVR typing despite their evident epidemiologic implication. These isolates had several nucleotide substitutions (n = 12) distributed throughout a 246-nucleotide region of the SVR, suggesting that the change was due to recombination rather than spontaneous mutation. The same underlying intragenomic recombination (Fig. 1) event between flaB and flaA (5-bp differences in a 150-nucleotide region) could also be shown in another isolate (D2692) with an unstable flaA during outbreak 3. Therefore, the epidemiologic concordance for all 33 confirmed outbreak isolates was 1.00 for flaB typing, whereas for flaA it was only 0.97. Furthermore, false-negative outbreak cross-classification results were not observed for MLST or for flaB. It was not possible to type C. coli by MLST (2), whereas the other typing methods were applicable without modification for the isolates tested (n = 4).
TABLE 3.
Cross-classification concordance results between PFGE and MLST, flaA, or flaB (C. jejuni, n = 42; all possible pairs, n = 861)
| PFGE | MLSTa
|
flaAb
|
flaBc
|
|||
|---|---|---|---|---|---|---|
| No. of matches | No. of mismatches | No. of matches | No. of mismatches | No. of matches | No. of mismatches | |
| Match | 48 | 0 | 36 | 12 | 48 | 0 |
| Mismatch | 52 | 761 | 33 | 780 | 36 | 777 |
Overall concordance with PFGE, 93.96%.
Overall concordance with PFGE, 94.77%.
Overall concordance with PFGE, 95.82%.
In conclusion, PFGE remains the most discriminatory typing method for Campylobacter. However, flaB typing is a rapid, reproducible, discriminatory, and stable screening tool. Our data demonstrate that flaB is more stable than flaA, probably due to less selective pressure. Larger studies of epidemiologically well-characterized isolates are needed to prove definitively the superiority of flaB for the monitoring of Campylobacter populations.
Acknowledgments
This work was supported by a grant from the Bundesministerium für Bildung und Forschung (BMBF, FKZ 01KI0204).
We thank U. Busch and A. Heiβenhuber, Bayerisches Landesamt für Gesundheit und Lebensmittelsicherheit (Oberschleiβheim, Germany), J. Bockemühl, Institut Hygiene und Umwelt (Hamburg, Germany), and J. Steigerwalt and P. Fields, Centers for Disease Control and Prevention (Atlanta, Ga.), for kindly providing us with Campylobacter strains. We are grateful to A. Lagemann, U. Keckevoet (Münster, Germany), and M. Müller (Berlin, Germany) for skillful technical assistance.
REFERENCES
- 1.Alm, R. A., P. Guerry, and T. J. Trust. 1993. Significance of duplicated flagellin genes in Campylobacter. J. Mol. Biol. 230:359-363. [DOI] [PubMed] [Google Scholar]
- 2.Dingle, K. E., F. M. Colles, D. R. A. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. L. Willems, R. Urwin, and M. C. J. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dingle, K. E., F. M. Colles, R. Ure, J. A. Wagenaar, B. Duim, F. J. Bolton, A. J. Fox, D. R. A. Wareing, and M. C. J. Maiden. 2002. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 8:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harmsen, D., H. Claus, W. Witte, J. Rothgänger, H. Claus, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrington, C. S., F. M. Thomson-Carter, and P. E. Carter. 1997. Evidence for recombination in the flagellin locus of Campylobacter jejuni: implications for the flagellin gene typing scheme. J. Clin. Microbiol. 35:2386-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meinersmann, R. J., L. O. Helsel, P. I. Fields, and K. L. Hiett. 1997. Discrimination of Campylobacter jejuni isolates by fla gene sequencing. J. Clin. Microbiol. 35:2810-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 9.Ribeiro, C. D., and J. A. Frost. 2000. Family clusters of Campylobacter infection. Commun. Dis. Public Health 3:274-276. [PubMed] [Google Scholar]
- 10.Ribot, E. M., C. Fitzgerald, K. Kubota, B. Swaminathan, and T. J. Barrett. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson, D. A., S. K. Hollingshead, J. M. Musser, A. J. Parkinson, D. E. Briles, and M. J. Crain. 1998. The IS1167 insertion sequence is a phylogenetically informative marker among isolates of serotype 6B Streptococcus pneumoniae. J. Mol. Evol. 47:222-229. [DOI] [PubMed] [Google Scholar]
- 12.Sails, A. D., B. Swaminathan, and P. I. Fields. 2003. Utility of multilocus sequence typing as an epidemiological tool for investigation of outbreaks of gastroenteritis caused by Campylobacter jejuni. J. Clin. Microbiol. 41:4733-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Struelens, M. J. 1996. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin. Microbiol. Infect. 2:2-11. [DOI] [PubMed] [Google Scholar]
- 14.Thurm, V., E. Dinger, O. Lyytikäinen, A. Wiebelitz, D. Lange, R. Fischer, H. Oppermann, and D. Mäde. 1999. Epidemiology of food-borne illnesses caused by Campylobacter infections. Bundesgesundheitsbl. Gesundheitsforsch. Gesundheitsschutz. 42:206-211. [Google Scholar]
- 15.Thurm, V., R. Stark, D. Mäde, S. Fanghänel, W. Berger, H. Knobloch, and D. Lange. 2000. Raw milk as a source of foodborne Campylobacter infections. Bundesgesundheitsbl. Gesundheitsforsch. Gesundheitsschutz. 43:777-780. [Google Scholar]
- 16.Wassenaar, T. M., B. N. Fry, and B. A. M. van der Zeijst. 1995. Variation of the flagellin gene locus of Campylobacter jejuni by recombination and horizontal gene transfer. Microbiology 141:95-101. [DOI] [PubMed] [Google Scholar]
- 17.Wassenaar, T. M., and D. G. Newell. 2000. Genotyping of Campylobacter spp. Appl. Environ. Microbiol. 66:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]