Fig. 1.
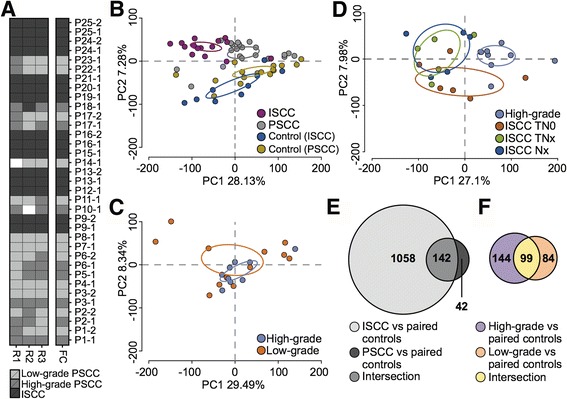
Sample classification, general variability of normalized gene expression and differential gene expression in the selected FFPE PSCC and ISCC samples. a Sample classification according to independent reviews by three expert histopathologists. Each of the first three columns in the heatmap corresponds to one of the three involved pathologists (R1, R2 and R3) and rows to the selected samples. The color of each heatmap cell indicates the corresponding sample classification (see legend, white = no review for technical reasons). The FC column represents the final classification based on two concordant reviews. b Plot shows the result of a Principal component analysis (PCA) of normalized gene expression data obtained from all the included samples. ISCC and PSCC samples are clearly separated. Ellipses delineate 95% confidence level for each sample class respectively. c PCA plot of normalized gene expression data from low- and high-grade PSCC. Samples are widespread and show no clear separation. d PCA plot of normalized gene expression data from ISCC and high-grade PSCC. TN0, TNx and Nx ISCC samples are widespread and do not show recognizable differences. High-grade PSCC are clearly separated from ISCC samples. e Venn diagram illustrate the numbers of differentially expressed genes in ISCC and PSCC, including the intersection between both. f The Venn diagram represents the numbers of differentially expressed genes in high-grade and low-grade PSCC, including the intersection between both
