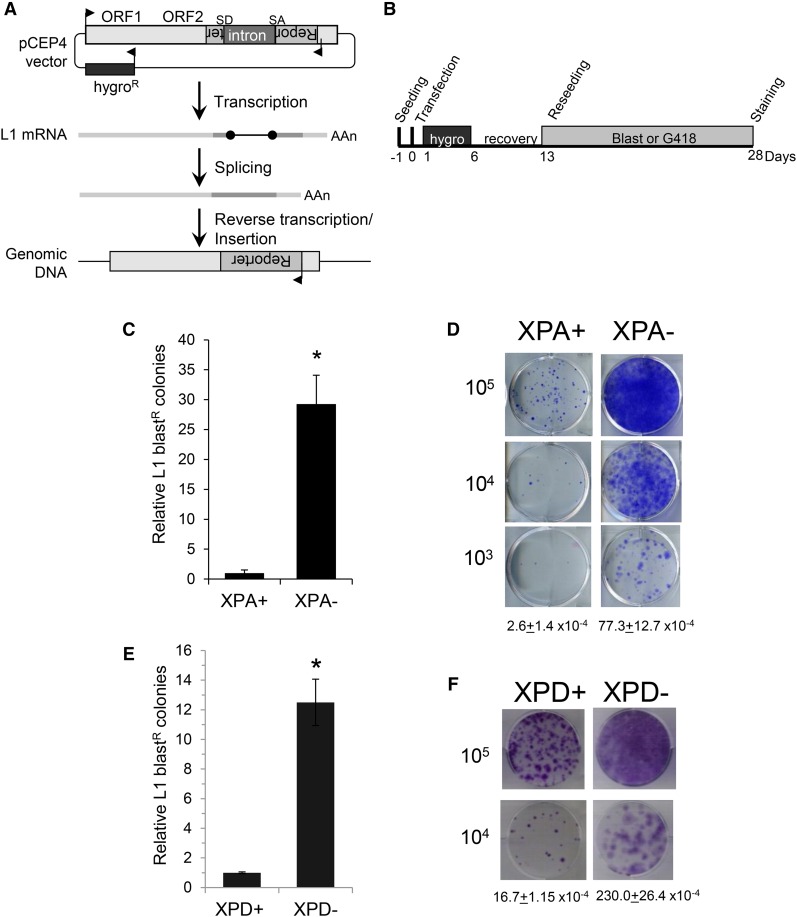
Figure 2.
XPD and XPA proteins limit L1 retrotransposition. (A) Schematic of the L1-retrotransposition assay. The L1 vector is a pCEP4 episomal vector carrying a hygromycin resistance (hygroR) gene, for the selection of transfected cells, and a full length L1 element tagged at the 3′ end with a retrotransposition cassette. The retrotransposition cassette consists of a reporter gene such as blasticidin resistance (blastR) or a neomycin resistance (neoR) gene interrupted with an intron. The reporter gene is in the reverse orientation in comparison to the L1 element and its transcription is driven by its own promoter, whereas the intron is in the sense orientation (direction of transcription is indicated by arrows). The splice donor and acceptor of the intron are indicated as SD and SA. The resistance gene becomes functional after L1 transcription, splicing, and TPRT of a new L1 copy into the genome. Therefore, only when retrotransposition of the cassette occurs can cells grow under selection conditions (blastR or neoR colonies). (B) Schematic of the protocol of the L1-retrotransposition assay used with the hygroR pCEP4 episomal vectors. The day after transfection, cells containing plasmid are selected with hygromycin for 3 days. Then, selection medium is removed and replaced by growth medium for recovery. Ten days post-transfection, cells were reseeded in six-well plates at serial dilution, from 106 to 103 cells, in triplicate and grown under the appropriate selection. Two weeks later, cells were fixed and stained in crystal violet solution and the number of blastR or neoR colonies in each well is counted. (C) Relative retrotransposition rates of a blastR-tagged L1 reporter element (TAM102/L1.3) were determined in an XPA-deficient cell line (XPA-) and a commercially available stably complemented XPA-deficient cell line (XPA+). The results were normalized relative to XPA+, which was arbitrarily set to 1.0. L1 retrotransposition assay was performed three times independently. Bars represent the average and SEM from the three independent experiments. Statistical significance is indicated by * P-value = 1 × 10−15 (two-tailed, two-sample t-test). (D) Representative example of blastR colony formation resulting from L1 retrotransposition assay performed in XPA+ and XPA- cells. The L1 retrotransposition rate for each cell line is indicated below. (E) Relative number of blastR colonies resulting from L1 retrotransposition assay performed in XPD+ and XPD- cell lines. The results were normalized relative to the XPD+, which was arbitrarily set to 1.0. L1 retrotransposition assay was performed three times independently. Bars represent the average with SEM from the three independent experiments. Statistical significance is indicated by * P-value = 6.5 × 10−6 (two-tailed, two-sample t-test). (F) Representative example of blastR colony formation resulting from L1 retrotransposition assay performed in XPD+ and XPD- cells. The L1 retrotransposition rate for each cell line is indicated below.