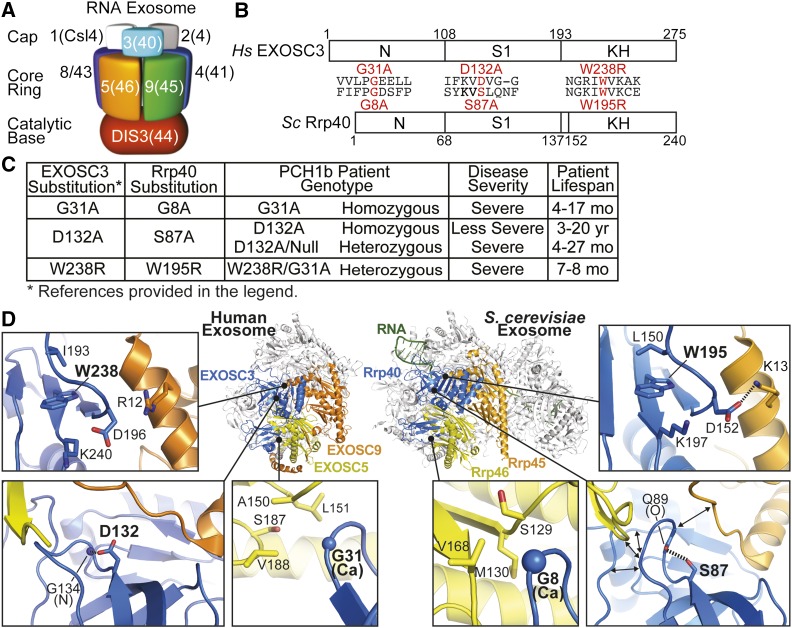
Figure 1.
Overview of PCH1b-associated amino acid substitutions in the human EXOSC3 subunit of the RNA exosome, and the corresponding substitutions in the budding yeast ortholog, Rrp40, examined in this study. (A) The RNA exosome is an evolutionarily conserved ribonuclease complex composed of 10 different subunits—three cap subunits: EXOSC1/Csl4 (Human/S. cerevisiae); EXOSC2/Rrp4; EXOSC3/Rrp40, six core subunits: EXOSC4/Rrp41; EXOSC5/Rrp46; EXOSC6/Mtr3; EXOSC7/Rrp42; EXOSC8/Rrp43; EXOSC9/Rrp45, and a catalytic base subunit: DIS3/Rrp44. Mutations in the EXOSC3 gene encoding a cap subunit [light blue, labeled with 3(40)] cause PCH1b (Wan et al. 2012), mutations in the EXOSC8 gene, encoding a core subunit [purple, labeled with 8(43)] cause PCH1c (Boczonadi et al. 2014), and mutations in the EXOSC2 gene, encoding a cap subunit [gray, labeled with 2(4)] cause a novel syndrome (Di Donato et al. 2016). (B) EXOSC3 and its budding yeast ortholog, Rrp40, contain three different domains: an N-terminal domain, a central S1 putative RNA binding domain, and a C-terminal putative RNA binding KH (K homology) domain. The position and flanking sequence of the major amino acid substitutions in PCH1b-associated human EXOSC3, and the substitutions generated in the S. cerevisiae ortholog of EXOSC3, Rrp40 (shown in red) are indicated. (C) Summary of the major amino acid substitutions identified in EXOSC3 in PCH1b patients (Wan et al. 2012) with corresponding substitutions in budding yeast Rrp40 generated in this study. Patient genotypes (Homozygous/Heterozygous) are reported, and disease severity and patient lifespans for each substitution (Wan et al. 2012; Schwabova et al. 2013; Eggens et al. 2014) are also presented to provide some context for the functional consequences of the different amino acid substitutions. (D) PCH1b-associated substitutions occur in EXOSC3 residues located near RNA exosome subunit interfaces. Structural models of the human RNA exosome (left) [PDB#2NN6 (Liu et al. 2006)] and S. cerevisiae RNA exosome (right) [PDB#4IFD; (Makino et al. 2013)] are depicted. The nine subunit human RNA exosome structure highlights the EXOSC3 (blue), EXOSC5 (yellow), and EXOSC9 (yellow) subunits. Zoomed-in views of three subunit interface regions show the locations of PCH1b-associated EXOSC3 substituted residues (G31, D132, and W238 residues in large, bold font). The S. cerevisiae RNA exosome structure highlights the orthologous Rrp40 (blue), Rrp46 (yellow), and Rrp45 (orange) subunits. Zoomed-in views show the locations of the evolutionarily conserved residues (G8, S87, and W195). S. cerevisiae Rrp40 G8 (EXOSC3 G31) is located in a hydrophobic pocket at an interface with Rrp46 (EXOSC5). Rrp40 S87 (EXOSC3 D132) and the backbone of Q89 are positioned to form a hydrogen bond that may organize the loop between strands β3 and β4, which is at the interface with both Rrp46 (EXOSC5) and Rrp45 (EXOSC9). Rrp40 W195 (EXOSC3 W238) sits in a large pocket, and could be important for positioning the loop of Rrp40 that forms an interface with Rrp45 (EXOSC5).