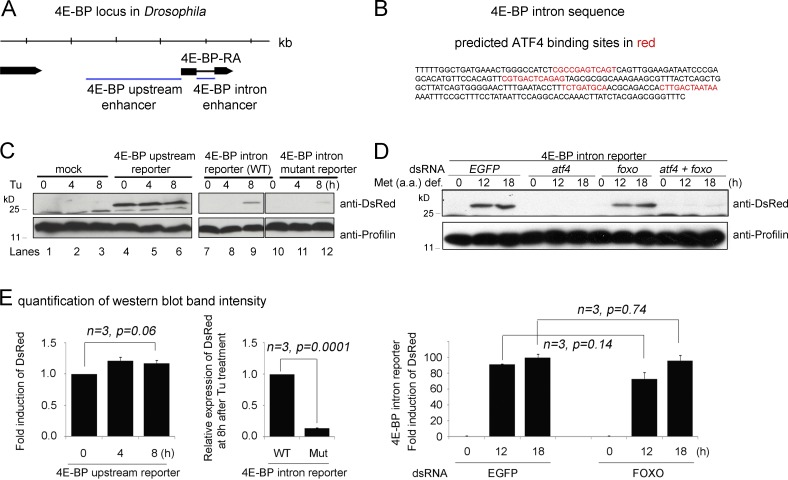
Figure 3.
Functional ATF4-binding sites are present in the 4E-BP intron. (A) A diagram of the 4E-BP (thor) genomic locus. Putative regulatory sequences (marked as blue lines) were subcloned upstream of dsRed to generate the upstream enhancer and the intron enhancer reporters. (B) The sequence of the intron enhancer. Predicted ATF4 binding sites are marked in red. (C) Upstream and intron (wild type [WT] and mutant for ATF4-binding sites) enhancer reporter expression in response to tunicamycin (Tu) treatment as shown by Western blot. The numbers above indicate hours of Tu treatment. The upstream enhancer-dsRed expression is shown in lanes 4–6, whereas the intron enhancer-dsRed is in lanes 7–9. The intron enhancer-dsRed with putative ATF4-binding sites mutated is shown in lanes 10–12. Antiprofilin blots are shown as controls (bottom). (D) 4E-BP intron reporter expression detected through Western blot after being cultured in a methionine-deficient media. The indicated genes (EGFP, atf4, and foxo) were knocked down before the analysis. The antiprofilin blot is shown as a control for those lanes (bottom). (E) Graphs showing quantified and normalized Western blot bands from gels in C and D. Error bars show SE. A t test was used to derive p-values.