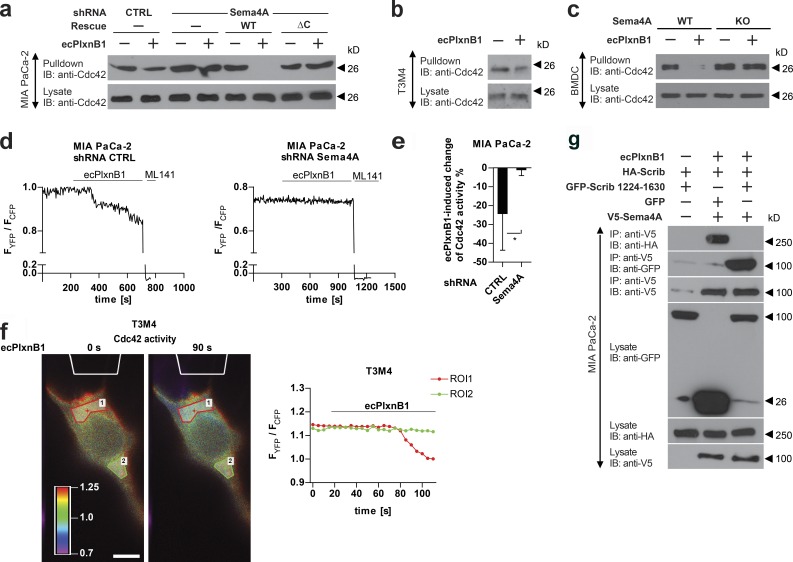
Figure 5.
Sema4A reverse signaling regulates the activity of Rac1 and Cdc42. (a) MIA PaCa-2 cells stably expressing control shRNA or Sema4A shRNA either alone or together with shRNA-resistant wild-type (WT) or mutated (ΔC) Sema4A (as indicated) were starved and stimulated with or without 150 nM ecPlxnB1, and active Cdc42 was precipitated as described in Materials and methods. (b) T3M4 cells were treated with or without 150 nM ecPlxnB1, and Cdc42 activity was measured (pulldown). (c) Mature BMDCs generated from wild-type or Sema4A knockout mice (Sema4A KO) were stimulated with or without 150 nM ecPlxnB1 for 20 min. Cells were then lysed and the activity of Cdc42 was analyzed (pulldown). (d and e) MIA PaCa-2 cells stably expressing control or Sema4A shRNA were transfected with a FRET biosensor for Cdc42 and FRET ratios in response to 150 nM ecPlxnB1, and 100 µM of the Cdc42 inhibitor ML141 were analyzed as described in Materials and methods. (d) Representative traces are shown. CTRL, control. (e) The ecPlxnB1-induced change of Cdc42 activity was calculated as a percentage of the difference between the FRET ratio at baseline (before treatments) and the FRET ratio after ML141 treatment. n = 12 for control shRNA, n = 8 for Sema4A shRNA. (f) ecPlxnB1 was applied to one side of a T3M4 cell through a micropipette (as outlined in white at the top of the images) as described in Materials and methods. Shown are representative images and the corresponding FRET ratios within two different areas of the cell over time. The regions of interest (ROIs) indicate regions proximal or distal to the site of ecPlxnB1 application, respectively. Bar, 10 µm. (g) MIA PaCa-2 cells were transfected with constructs encoding HA-Scrib, V5-Sema5A, and the GFP-tagged C-terminal portion of Scrib (GFP-Scrib amino acids 1224–1630) as indicated. Cells were stimulated with or without 150 nM ecPlxnB1. Proteins interacting with Sema4A were immunoprecipitated using V5 antibody and visualized using respective antibodies. IB, immunoblotting; IP, immunoprecipitation. Error bars represent means ± SD. *, P < 0.05.