Abstract
We characterized the single-nucleotide polymorphisms in the rRNA operon and variable numbers of tandem repeats in the lipoprotein gene MG309 among Mycoplasma genitalium strains from clinical specimens by PCR and sequencing. Analysis of 31 M. genitalium-infected patient specimens and 7 American Type Culture Collection strains identified six types of rRNA sequences and 11 different numbers of MG309 repeats. Examination of sequential specimens from 10 patients showed that these genotypes were stable for at least 5 weeks. These data suggest the potential usefulness of the rRNA genotypes and the MG309 repeats for genotyping of M. genitalium.
The increasing importance of Mycoplasma genitalium as a human pathogen (4, 7-9) creates a need to develop typing methods for a better understanding of its epidemiology and pathogenesis. Presently, neither phenotypic nor serologic typing methods are available for M. genitalium, due largely to the difficulty in culturing this organism. While molecular typing of M. genitalium has been attempted by one group by using amplified-fragment length polymorphism (5), this method requires purified DNA from cultured strains and is, therefore, not applicable to typing directly from clinical specimens. Other approaches are clearly needed. The availability of the whole genome sequence of M. genitalium (3) affords the opportunity to search for genetic markers for typing purposes. The primary goal of this study was to characterize single-nucleotide polymorphisms in the rRNA operon and variable numbers of short tandem repeats (STR) in a putative lipoprotein gene (MG309) among M. genitalium clinical specimens as possible ways for genotyping this organism.
M. genitalium specimens.
Seven M. genitalium strains (ATCC 33530, 49123, 49895, 49896, 49897, 49898, and 49899) were obtained from the American Type Culture Collection (ATCC; Manassas, Va.) and were cultured in SP-4 medium according to the ATCC instructions. The strain ATCC 33530 is also known as G37 and was used to determine the M. genitalium genome sequence (3). First, voided urine samples were obtained between 2002 and 2003 from 31 men with acute urethritis attending the New Orleans Delgado sexually transmitted disease clinic. An additional one or two follow-up urine specimens were collected from 10 of these men at intervals ranging from 10 to 36 days. All urine samples were positive for M. genitalium by our PCR assay as described elsewhere (8). Informed consent was obtained from patients, and study protocols were approved by the Louisiana State University Health Sciences Center Institutional Review Board. Genomic DNA in all specimens was extracted as previously described (8).
PCR and sequencing.
Based on the genome sequence of M. genitalium strain G37 (3), primers were designed to amplify two fragments (16S-ITS1-23S and 23S-ITS2-5S) of the rRNA operon (Fig. 1) and the STR region of the putative lipoprotein gene MG309 (Fig. 2). To obtain PCR products in sufficient quality and quantity for sequencing, we performed a primary PCR for all samples and a secondary nested PCR for those samples for which the initial PCR products were not visible or were very faint on agarose gels. To amplify the fragment 16S-ITS1-23S, the primers were Mg16-1240F (5′-CAATGGCCAATACAAACAGTAGCCAA-3′) and Mg23-2033R (5′-GCAAGCCCTACAACCCCTATCCTCA-3′) for the primary PCR and Mg16-1381F (5′-CCCGTCAAACTATGAAAGCTGGTA-3′) and Mg23-1926R (5′-AGATGTTTCACTTCACCACGTATC-3′) for the nested PCR. To amplify the fragment 23S-ITS2-5S, the primers were Mg23-4344F (5′-TCCCTATCTATTGTGCCCAC-3′) and Mg5-4899R (5′-GAGCGTATACTTAGTCAAAGGTTG-3′) for the primary PCR and Mg23-4380F (5′-TTTGCTTCTAGTACGAGAGGACCGG-3′) and Mg5-4779R (5′-CTTACCTATTCTCAGTATTACTAC-3′) for the nested PCR. To amplify the MG309 STR region, the primers were 384275F (5′-GTGCTAGAGAAGTGTTTCTAGGATC-3′) and 384655R (5′-AACTAGCAGAACGTAACCAACC-3′) for the primary PCR and 384330F (5′-TTGTGAATATTGCGGTGAGG-3′) and 384655R for the seminested PCR. In the primary PCR, 0.5 or 2 μl of DNA extract was used in a final volume of 20 μl containing 0.5 μM each primer, 0.2 mM deoxynucleoside triphosphates, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2.5 mM MgCl2, and 0.5 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer, Foster City, Calif.). In the secondary PCR, 0.5 to 2 μl of the primary PCR products was used in a final volume of 50 μl. The first round of PCR was performed with a touchdown protocol as described elsewhere (6). For the second round of PCR, the cycling parameters were 1 cycle of 95°C for 9 min followed by 30 cycles of 95°C for 45 s, 50°C for 1 min, and 72°C for 2 min. All PCR products were purified by use of the DNA Clean and Concentrator 5 kit (Zymo Research, Orange, Calif.) and were sequenced directly. In addition, eight PCR products were also sequenced after subcloning by use of the TA Cloning Kit (Invitrogen, San Diego, Calif.). DNA sequencing was carried out by use of an ABI PRISM 3100 automated capillary sequencer (Perkin-Elmer). Nucleic acid sequences were analyzed by use of the MultAlin software (2). The sequence of the genome of M. genitalium strain G37 (3) was chosen as the prototype sequence, and all sequences obtained in this study were compared with this sequence.
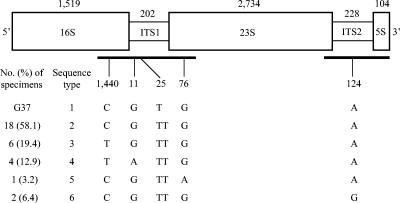
FIG. 1.
Schematic representation and sequence variation of the M. genitalium rRNA operon. ITS1 and ITS2 refer to the internal transcribed spacers between the 16S and 23S rRNA genes and between the 23S and 5S rRNA genes, respectively. The size (in base pairs) indicated above each region was derived from the M. genitalium genome sequence (GenBank accession no. NC_000908). The line length is not proportional to the number of nucleotides involved. The black bars represent the PCR-amplified fragments, 16S-ITS1-23S (approximately 800 bp) and 23S-ITS2-5S (approximately 450 bp). The lines leading downwards from the fragment point to the position of each polymorphic nucleotide. The TT change at position 25 in ITS1 represents two Ts compared to the single T in the G37 prototype strain. Numbering starts from the first nucleotide for each region. Only the first sample is included for patients who had sequential samples. The rRNA sequences of all seven ATCC strains matched the G37 sequence exactly.
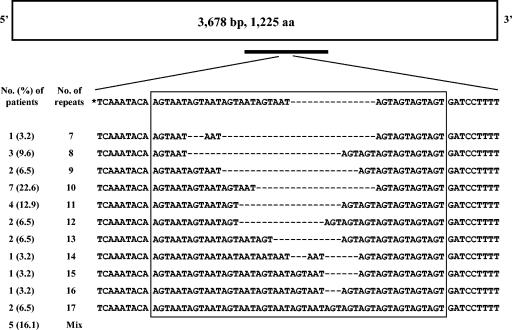
FIG. 2.
Schematic representation and STR of the putative lipoprotein gene MG309 of M. genitalium in 31 patient specimens. The numbers of nucleotides and deduced amino acids (aa) in the coding region are indicated. The black bar represents the fragment amplified by PCR (approximately 350 bp). The nucleotide alignment includes only a small portion of the amplified region and only one representative sequence for different repeat patterns. The sequence on the top (*) was derived from the M. genitalium genome sequence (GenBank accession no. NC_000908). The boxed regions represent the trinucleotide repeats (AGT or AAT). Hyphens indicate gaps introduced to optimize alignment. Specimens with identical repeat numbers may have sequence variation between repeat units (data not shown). The sequences for the five patients containing a mixture (Mix) of ≥2 repeat patterns are not shown.
Observations and interpretations. Analysis of the 16S-ITS1-23S rRNA fragment in 31 first-visit patient specimens and seven ATCC strains identified four unique sequences that varied from the prototype at four nucleotide positions (Fig. 1). The C-to-T nucleotide change in the 16S rRNA gene observed in 10 of our strains was identical to that found in Danish culture strains (GenBank accession no. AY466443). The 23S-ITS2-5S fragment showed sequence variation at a single position in the ITS2 region compared to that of the prototype (Fig. 1). Combination of the sequence polymorphisms of the 16S-ITS1-23S and 23S-ITS2-5S fragments resulted in six different genotypes (Fig. 1). All seven ATCC strains were identical (type 1 or prototype). The nucleotide changes for rRNA types 2 to 6 were verified by repeating DNA extraction from a different aliquot of the original samples followed by PCR and direct sequencing.
The MG309 STR was highly variable in repeat numbers (7 to 17) among 31 different patients, with 10 and 11 repeats being the most common (Fig. 2). In addition, the distribution patterns of the repeat units (AGT and AAT) varied between different patients (for an example, see GenBank accession nos. AY386810, AY644507, and AY644508). All seven ATCC strains had the same repeat number and distribution pattern compared to that of the published sequence for the G37 strain (3). For five patients, direct sequencing of the PCR products showed a mixture of two or more different MG309 sequences. By subcloning the PCR products and subsequent sequencing of individual clones, we verified the presence of a mixture of two or three MG309 sequence patterns in all five patients (data not shown). To rule out the possibility of sample cross-contamination, we repeated DNA extraction from a different aliquot of the original samples for these five patients followed by PCR and DNA sequencing and obtained consistent results. The presence of mixed sequences in the same patient suggests that the patient is coinfected with two or more different strains or that mutation within the MG309 STR occurs relatively frequently in some M. genitalium strains.
To examine the stability of the rRNA genotypes and MG309 STR, two or three sequential specimens were obtained from each of 10 patients at an interval of 10 to 36 days between specimens. No change of rRNA genotypes or MG309 repeat numbers was observed in nine of those patients (including two with mixed MG309 sequences). Only one patient showed a change from a mixture of 10, 11, and 12 repeats to a mixture of 11 and 12 repeats. This finding, together with the fact that the G37 strain presently propagated in our laboratory has the same rRNA and MG309 sequences as the old G37 strain used a decade ago to determine the M. genitalium genome sequence (3), suggests a temporal stability of these sequences for at least 5 weeks.
The variability of the MG309 repeats is much greater than that of the rRNA sequences (Table 1), suggesting that the former may be evolving more rapidly than the latter. Notably, MG309 repeat number variations are considerable within each rRNA type. The discriminatory power of MG309 STR is further improved by the variation of the distribution patterns of the two types of repeat units among different patients who had an identical number of MG309 repeats and who had an identical rRNA genotype (data not shown). Given the different degree of variability in the rRNA sequences and MG309 STR, a combination of these markers should be more useful than a single marker for studying sexual transmission patterns of M. genitalium. Further studies are needed of the stability of these genetic markers, particularly MG309, over longer periods of time.
TABLE 1.
Comparison of the rRNA sequence types and MG309 repeats in seven ATCC strains and 31 patient specimens
| rRNA genotype | No. of specimens with MG309 repeat no.:
|
No. of specimens with a mixture of repeats | Total | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | |||
| 1 | 7a | 7 | |||||||||||
| 2 | 1 | 1 | 1 | 3 | 1 | 2 | 1 | 1 | 1 | 2 | 4 | 18 | |
| 3 | 1 | 2 | 3 | 6 | |||||||||
| 4 | 1 | 1 | 1 | 1 | 4 | ||||||||
| 5 | 1 | 1 | |||||||||||
| 6 | 1 | 1 | 2 | ||||||||||
| Total | 1 | 3 | 2 | 7 | 4 | 9 | 2 | 1 | 1 | 1 | 2 | 5 | 38 |
ATCC strains.
Given the diversity we observed among our clinical specimens, the fact that all seven ATCC strains we evaluated are identical in both the rRNA genotypes and MG309 STR is surprising. However, our results are consistent with those of Kokotovic et al. (5), who found that these seven ATCC strains had the same DNA fingerprinting profile. The ATCC organisms were initially isolated from a variety of body sites in different patients over two decades (1, 10, 11). The reason for lack of genetic differences in these strains is unclear. It could reflect the fact that only one M. genitalium strain is cultivable, or it could be that these isolates represent culture contamination by the original G37 strain. In any case, investigators should be aware of the fact that the ATCC M. genitalium strain collection does not reflect the potential genetic variability of clinical strains.
Nucleotide sequence accession numbers.
Representative nucleotide sequences obtained in this study have been deposited in GenBank under accession numbers AY374418 to AY374422 for rRNA genotypes 2 to 6, respectively, and AY386807 to AY386817 for 11 different patterns of MG309 repeats.
Acknowledgments
This work was supported by the Louisiana Board of Regents grant HEF (2001-2004)-04.
We thank Mary Welch and Judy Burnett for technical assistance.
REFERENCES
- 1.Baseman, J. B., S. F. Dallo, J. G. Tully, and D. L. Rose. 1988. Isolation and characterization of Mycoplasma genitalium strains from the human respiratory tract. J. Clin. Microbiol. 26:2266-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, et al. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 4.Jensen, J. S., R. Orsum, B. Dohn, S. Uldum, A. M. Worm, and K. Lind. 1993. Mycoplasma genitalium: a cause of male urethritis? Genitourin. Med. 69:265-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kokotovic, B., N. F. Friis, J. S. Jensen, and P. Ahrens. 1999. Amplified fragment length polymorphism fingerprinting of Mycoplasma species. J. Clin. Microbiol. 37:3300-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma, L., G. Kutty, Q. Jia, and J. A. Kovacs. 2003. Characterization of variants of the gene encoding the p55 antigen in Pneumocystis from rats and mice. J. Med. Microbiol. 52:955-960. [DOI] [PubMed] [Google Scholar]
- 7.Manhart, L. E., C. W. Critchlow, K. K. Holmes, S. M. Dutro, D. A. Eschenbach, C. E. Stevens, and P. A. Totten. 2003. Mucopurulent cervicitis and Mycoplasma genitalium. J. Infect. Dis. 187:650-657. [DOI] [PubMed] [Google Scholar]
- 8.Mena, L., X. Wang, T. F. Mroczkowski, and D. H. Martin. 2002. Mycoplasma genitalium infections in asymptomatic men and men with urethritis attending a sexually transmitted diseases clinic in New Orleans. Clin. Infect. Dis. 35:1167-1173. [DOI] [PubMed] [Google Scholar]
- 9.Totten, P. A., M. A. Schwartz, K. E. Sjostrom, G. E. Kenny, H. H. Handsfield, J. B. Weiss, and W. L. Whittington. 2001. Association of Mycoplasma genitalium with nongonococcal urethritis in heterosexual men. J. Infect. Dis. 183:269-276. [DOI] [PubMed] [Google Scholar]
- 10.Tully, J. G., D. L. Rose, J. B. Baseman, S. F. Dallo, A. L. Lazzell, and C. P. Davis. 1995. Mycoplasma pneumoniae and Mycoplasma genitalium mixture in synovial fluid isolate. J. Clin. Microbiol. 33:1851-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tully, J. G., D. Taylor-Robinson, R. M. Cole, and D. L. Rose. 1981. A newly discovered mycoplasma in the human urogenital tract. Lancet i:1288-1291. [DOI] [PubMed] [Google Scholar]