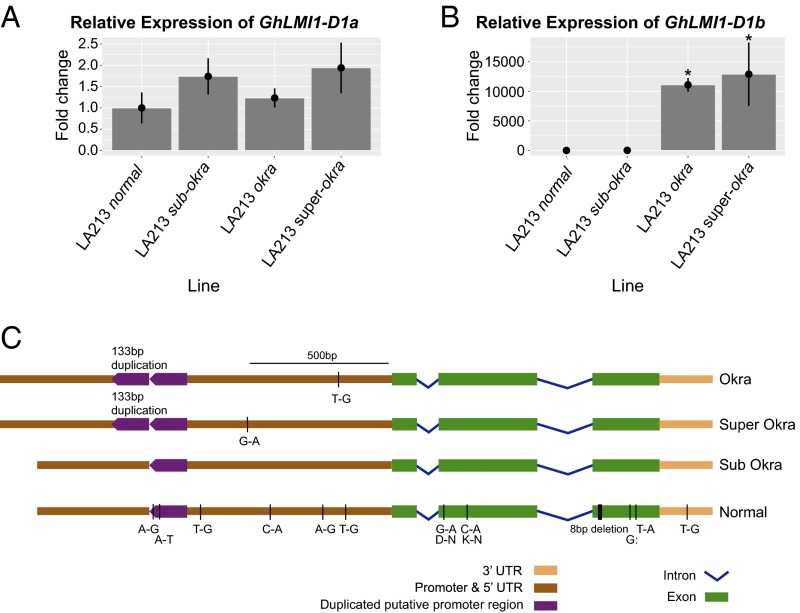
Fig. 3.
Nucleotide polymorphisms of GhLMI1-D1b gene and expression analysis of candidate genes using qRT-PCR among different leaf shapes. (A and B) Relative transcript levels of leaf shape candidate genes (GhLMI1-D1a and GhLMI1-D1b) among isolines (n = 3) at ∼90 d after planting. There is no difference in the expression of GhLMI1-D1a among leaf shapes but a significant increase of GhLMI1-D1b expression in okra and superokra. Error bars represent the SD of the fold change. (B) Asterisks represent statistically significant differences as determined by unpaired t tests at P < 0.05. (C) Nucleotide polymorphisms of GhLMI1-D1b gene among four major leaf shapes of tetraploid cotton. The 133-bp tandem duplicated region in the putative promoter was found only in okra and superokra and in parallel with elevated gene expression. The 8-bp deletion was found only in normal and causes a frameshift mutation and premature stop codon. All other polymorphisms are SNPs with unknown effect on the gene expression and protein function. Subokra is set as the standard to which the other three leaf shapes are compared.