Fig. 4.
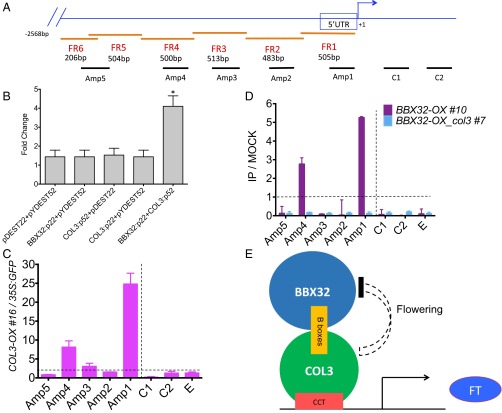
COL3 targets FT to regulate flowering. (A) Schematic representation of the FT gene and amplified promoter regions used for yeast and ChIP assays. FR (1–6) represent fragments cloned in yeast and Amp (1–5) represent amplicons amplified for ChIP assay. C1 and C2 represent regions of locus-control used in ChIP as described (10). (B) β-galactosidase activity in yeast FR1. The fold change of β-galactosidase activity of each construct was calculated by normalizing over control vectors. Bars represent the average of two biological replicates, and error bars represent the SEM. **P < 0.001 (unpaired t test). (C and D) Chromatin immunoprecipitation (ChIP) analysis of GFP-tagged COL3-OX #16 and Flag-tagged BBX32-OX #10 on the FT promoter. The 10-d-old GFP-tagged COL3-OX #16, 35S:GFP, Flag-tagged BBX32-OX #10, and Flag-tagged BBX32 OX_col3 #7 seedlings were grown in 16L:8D cycle at 22 °C and harvested at 2–3 h after dawn. The ratio of specific enrichment in the GFP-tagged COL3-OX #16 samples and that in the 35S:GFP sample is represented (C) whereas FLAG-tagged BBX32-OX #10; FLAG-tagged BBX32 OX_col3 #7 and that in mock (no antibody) (D) in each region were plotted. Experiments were independently repeated three times, each time with two biological replicates. C1 and C2 are the control regions and E (UBQ10) is a nonlocus control as described (10). Dotted line represents no enrichment. (E) Model representing mechanism of action of BBX32 in regulating flowering.