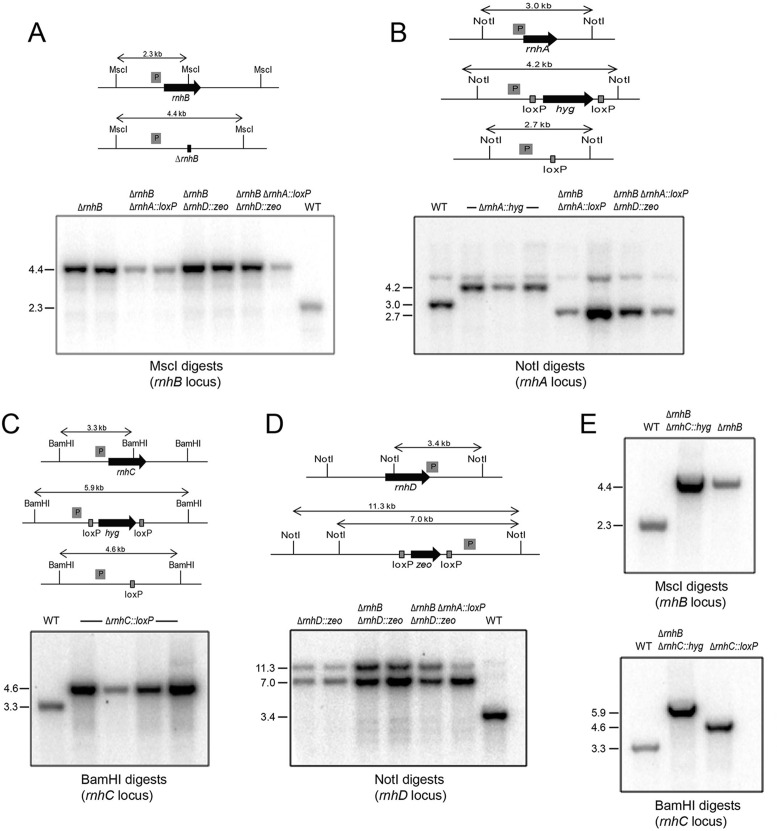
Figure 6.
Genotyping of Δrnh mutants. For each panel, schematic representations of the wild type and Δrnh genetic loci are shown with probe location (marked by a grey box ‘P’) and restriction endonuclease sites. Predicted hybridization products are indicated for wild type and deletion alleles. Below these schematics are autoradiograms of Southern blots of restriction endonuclease-digested chromosomal DNA hybridized with the indicated radiolabelled DNA probe from candidate recombinant strains in either wild type or Δrnh backgrounds, as indicated above each lane, along with chromosomal DNA derived from wild type M. smegmatis (WT). (A) Deletion of rnhB in WT (strain Mgm4085), ΔrnhA (strain Mgm4088), ΔrnhD (strain Mgm4090) and ΔrnhA ΔrnhD (strain Mgm4091). (B) Deletion of rnhA in WT (strain Mgm4087), ΔrnhB (strain Mgm4088), and ΔrnhB ΔrnhD (strain Mgm4091). (C) Deletion of rnhC in WT background after removal of the hygR marker via Cre recombinase (strain Mgm4084). (D) Deletion of rnhD in WT (strain Mgm4089), ΔrnhB (strain Mgm4090), ΔrnhA ΔrnhB (strain Mgm4091). (E) Confirmation of the ΔrnhB ΔrnhC double mutant (Mgm4086). Deletion of rnhB in the ΔrnhC background (top panel, see A for schematic) and rnhC in the ΔrnhB background (see C for schematic). In both top and bottom panels, the single mutants ΔrnhB (strain Mgm4085) and ΔrnhC::loxP (strain Mgm4084) were used as the controls in the last lanes, respectively.